The gene editing technique CRISPR/Cas9 has allowed researchers to make precise and impactful changes to an organism's DNA to fix mutations that cause genetic disease.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...

Martina Hrabinová's curator insight,
March 31, 2016 5:14 AM
A new way to avoid genotoxicity? Several types of CRISPR–Cas adaptive immune system have been described, each with a unique mechanism and protein composition. DNA cleavage by type III-A CRISPR–Cas systems relies on a complex known as Cas10–Csm and requires transcription of the target DNA. Type III-A CRISPR–Cas systems are also capable of degrading RNA, although why they should have an RNase function is not known. Jiang et al. now show that some phage targets can delay the transcription-dependent DNA cleavage mechanism of type III-A CRISPR–Cas systems but that CRISPR RNases prevent the replication of these phages. |

Hun-Joong Kim's curator insight,
March 29, 2016 10:30 PM
For first time, scientists use CRISPR-Cas9 to target RNA in live cells. They show that nuclear-localized RNA-targeting Cas9 (RCas9) is exported to the cytoplasm only in the presence of sgRNAs targeting mRNA and observe accumulation ofACTB, CCNA2, and TFRC mRNAs in RNA granules that correlate with fluorescence in situ hybridization. They also demonstrate time-resolved measurements of ACTB mRNA trafficking to stress granules. Their results establish RCas9 as a means to track RNA in living cells in a programmable manner without genetically encoded tags.

Eric Vincill's curator insight,
March 16, 2016 2:53 PM
Silas et al. discover that certain classes of the Cas1gene are fused to a reverse transcriptase gene (RT-Cas1) (see the Perspective by Sontheimer and Marraffini). These RT-Cas1 proteins are able to capture and directly incorporate both DNA and RNA into CRISPR loci. RT-Cas1 systems could be effective against parasitic RNA species, or even to modulate bacterial gene expression. |








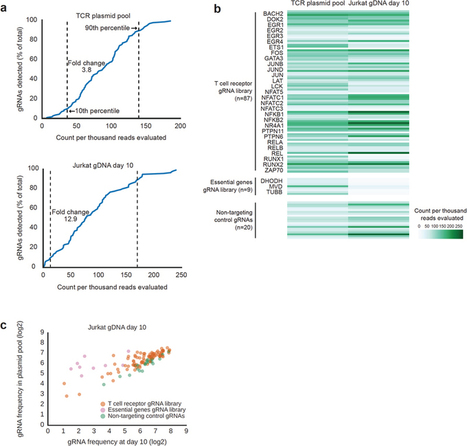







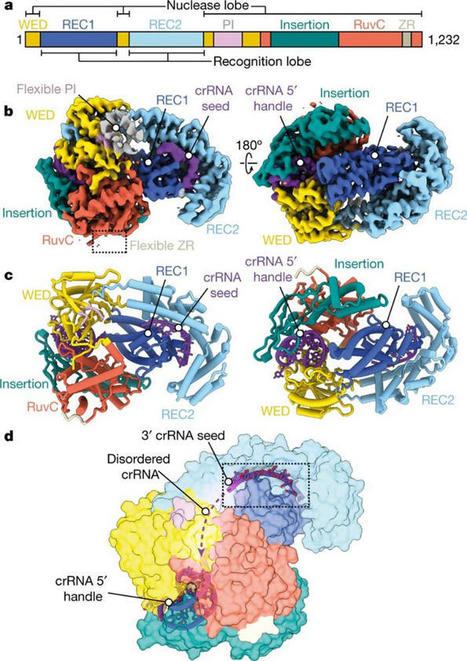


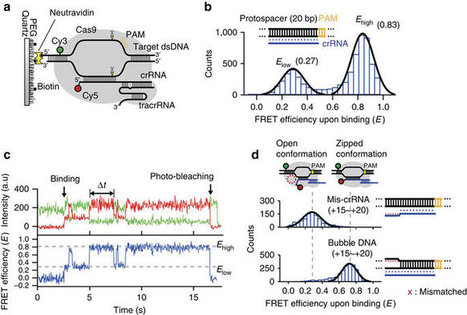









CRISPR/Cas9 method can lead to unintended DNA mutations that can have negative effects. Recently, Japanese researchers have developed a new gene-editing technique that is as effective as CRISPR/Cas9, yet significantly reduces these unintended mutations. In a new study published in Nature Communications , researchers led by Osaka University have introduced a new technique called NICER, based on the creation of several small cuts in single DNA strands by an enzyme Cas 9 nickase. For their first experiments, the research team used human lymphoblastic cells with a known heterozygous mutation in a gene called TK1. When these cells were treated with nickase to induce a single cut in the TK1 region, TK1 activity was recovered at a low rate. However, when nickase induced multiple cuts in this region on both homologous chromosomes, the efficiency of gene correction was increased approximately seventeen-fold via activation of a cellular repair mechanism. Because the NICER method does not involve DNA double-strand breaks or the use of exogenous DNA, this technique appears to be a safe alternative to conventional CRISPR/Cas9 methods.