Current approaches for inserting autonomous transgenes into the genome, such as CRISPR–Cas9 or virus-based strategies, have limitations including low efficiency and high risk of untargeted genome mutagenesis. Here, we describe precise RNA-mediated insertion of transgenes (PRINT), an approach for site-specifically primed reverse transcription that directs transgene synthesis directly into the genome at a multicopy safe-harbor locus. PRINT uses delivery of two in vitro transcribed RNAs: messenger RNA encoding avian R2 retroelement-protein and template RNA encoding a transgene of length validated up to 4 kb. The R2 protein coordinately recognizes the target site, nicks one strand at a precise location and primes complementary DNA synthesis for stable transgene insertion. With a cultured human primary cell line, over 50% of cells can gain several 2 kb transgenes, of which more than 50% are full-length. PRINT advantages include no extragenomic DNA, limiting risk of deleterious mutagenesis and innate immune responses, and the relatively low cost, rapid production and scalability of RNA-only delivery. Transgenes are inserted into human cells by 2-RNA delivery of a retroelement protein and template.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|












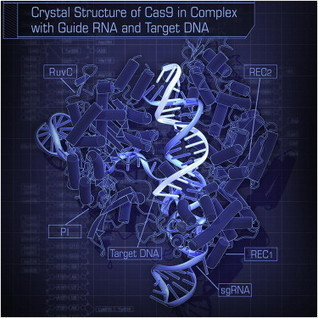



The recent approval of a CRISPR-Cas9 therapy for sickle-cell anemia demonstrates that gene-editing tools can do an excellent job of eliminating genes to cure inherited diseases. But it is still not possible to insert entire genes into the human genome to replace them with defective or deleterious genes. A new technique, called RNA-mediated Precise Transgene Insertion, or PRINT, exploits the ability of certain retrotransposons to efficiently insert whole genes into the genome without affecting other genome functions. PRINT would complement the recognized ability of CRISPR-Cas technology to deactivate genes, perform point mutations and insert short segments of DNA. For PRINT, one piece of delivered RNA encodes a common retroelement protein called the R2 protein, which has several active parts, including a nickase and a reverse transcriptase. The other RNA is the template for the transgenic DNA to be inserted, as well as the elements controlling gene expression