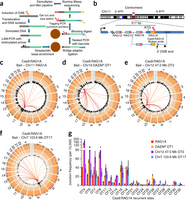
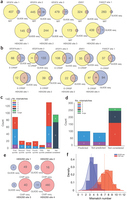
Fluorescence resonance energy transfer (FRET) reporters are commonly used in the final stages of nucleic acid amplification tests to indicate the presence of nucleic acid targets, where fluorescence is restored by nucleases that cleave the FRET reporters. However, the need for dual labelling and purification during manufacturing contributes to the high cost of FRET reporters. Here we demonstrate a low-cost silver nanocluster reporter that does not rely on FRET as the on/off switching mechanism, but rather on a cluster transformation process that leads to fluorescence color change upon nuclease digestion. Notably, a 90 nm red shift in emission is observed upon reporter cleavage, a result unattainable by a simple donor-quencher FRET reporter. Electrospray ionization–mass spectrometry results suggest that the stoichiometric change of the silver nanoclusters from Ag13 (in the intact DNA host) to Ag10 (in the fragments) is probably responsible for the emission colour change observed after reporter digestion. Our results demonstrate that DNA-templated silver nanocluster probes can be versatile reporters for detecting nuclease activities and provide insights into the interactions between nucleases and metallo-DNA nanomaterials. Here the authors present a non-FRET DNA-templated silver nanocluster probe that exhibits a distinct colour switch from green to red upon nuclease digestion, visible under UV excitation, offering a low-cost, effective alternative to fluorescent reporters for detecting nuclease activities.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|










A new tool could reduce the cost of diagnosing infectious diseases. Researchers have developed a new, less expensive means of detecting nuclease digestion, one of the critical steps in many nucleic acid detection applications, such as those used to identify COVID-19 and other infectious diseases. A new study published in the journal Nature Nanotechnology shows that this inexpensive tool, called Subak, is effective in determining when nucleic acid cleavage occurs, which happens when an enzyme called nuclease breaks down nucleic acids, such as DNA or RNA, into smaller fragments. The traditional method for identifying nuclease activity, the Fluorescence Resonance Energy Transfer (FRET) probe, is 62 times more expensive to produce than the Subak reporter.