Since its breakthrough development more than a decade ago, CRISPR has revolutionized DNA editing across a broad range of fields.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|




















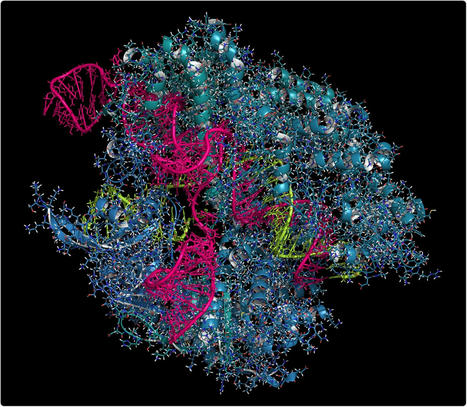








Researchers have developed a new genetic system to test and analyze the mechanisms underlying the results of CRISPR-based DNA repair. As described in Nature Communications, they have developed a sequence analyzer to help track on- and off-target mutational changes and how they are inherited from one generation to the next. The tool, called Integrated Classifier Pipeline (ICP), can reveal specific categories of mutations resulting from CRISPR editing. Developed in flies and mosquitoes, ICP provides a "fingerprint" of how genetic material is inherited, enabling scientists to track the source of mutational changes and the associated risks emerging from potentially problematic modifications. PKI can help unravel the complex biological issues that arise when determining the mechanisms behind CRISPR. Although developed in insects, ICP has great potential for human applications.