Your new post is loading...
 Your new post is loading...
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA generally becomes undetectable in upper airways after a few days or weeks postinfection. Here we used a model of viral infection in macaques to address whether SARS-CoV-2 persists in the body and which mechanisms regulate its persistence. Replication-competent virus was detected in bronchioalveolar lavage (BAL) macrophages beyond 6 months postinfection. Viral propagation in BAL macrophages occurred from cell to cell and was inhibited by interferon-γ (IFN-γ). IFN-γ production was strongest in BAL NKG2r+CD8+ T cells and NKG2Alo natural killer (NK) cells and was further increased in NKG2Alo NK cells after spike protein stimulation. However, IFN-γ production was impaired in NK cells from macaques with persisting virus. Moreover, IFN-γ also enhanced the expression of major histocompatibility complex (MHC)-E on BAL macrophages, possibly inhibiting NK cell-mediated killing. Macaques with less persisting virus mounted adaptive NK cells that escaped the MHC-E-dependent inhibition. Our findings reveal an interplay between NK cells and macrophages that regulated SARS-CoV-2 persistence in macrophages and was mediated by IFN-γ. Huot et al. show that interferon-γ (IFN-γ) regulates the persistence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in bronchoalveolar macrophages from cynomolgus macaques up to 18 months postinfection. Published in Nat. Immunology (Nov. 2, 2023): https://doi.org/10.1038/s41590-023-01661-4
Via Juan Lama
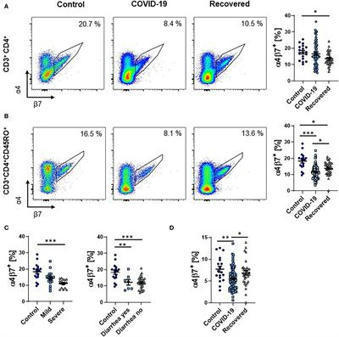
BackgroundInfection with the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes a wide range of symptoms including gastrointestinal manifestations, and intestinal epithelial cells are a target of the virus. However, it is unknown how the intestinal immune system contributes to systemic immune responses in coronavirus disease 2019 (COVID-19).MethodsWe characterized peripheral blood lymphocytes from patients with active COVID-19 and convalescent patients as well as healthy controls by flow cytometry.ResultsThe frequency and absolute number of circulating memory T and B cells expressing the gut homing integrin α4β7 integrin was reduced during COVID-19, whether gastrointestinal symptoms were present or not. While total IgA-expressing B cells were increased, gut-imprinted B cells with IgA expression were stable.ConclusionCOVID-19 is associated with a decrease in circulating adaptive immune cells expressing the key gut homing marker α4β7 suggesting that these cells are preferentially recruited to extra-intestinal tissues independently of α4β7 or that the systemic immune response against SARS-CoV-2 is at least numerically dominated by extraintestinal, particularly pulmonary, immune cell priming.
Rokote Laboratories are seeking investment for the vaccine and are already in discussions about clinical trials although no date has been set.
Compared with nasal swabs, saliva tests may better reflect infection deep in the lungs. To the known risk factors for developing severe COVID-19—age, male sex, or any of a series of underlying conditions—a new study adds one more: high levels of the virus in your saliva. Standard COVID-19 tests sample the nasal passage. But several new tests look for SARS-CoV-2, the pandemic coronavirus, in saliva, and the new work finds a striking correlation between high virus levels there and later hospitalization or death. If the results are confirmed, saliva tests could help doctors prioritize which patients in the early stages of the disease should receive medicines that drive down levels of the virus. “I thought it was pretty striking,” says Shane Crotty, a virologist at the La Jolla Institute for Immunology, who was not involved with the research. Crotty notes the results suggest virus levels in saliva reflect viral load deep in the lungs, where the disease does much of its damage in severe cases. “That is a fundamentally valuable insight,” Crotty says. The new work isn’t the first to link the body’s coronavirus load and disease outcome. Several research groups have found a correlation between high viral levels in the nasal passages at the time of a patient’s hospital admission and ultimate disease severity. But other groups have failed to find that same link. The standard test to detect SARS-CoV-2 samples nasal mucus using nasopharyngeal (NP) swabs. The procedure is unpleasant, but it is the customary way to sample respiratory pathogens. In recent months, however, several research groups have developed and received emergency use authorization from the U.S. Food and Drug Administration for tests detecting SARS-CoV-2 in saliva. Yale University researchers were among the first, and the university’s hospitals have been using both saliva and NP swab tests. In both cases, labs analyze the samples using quantitative reverse transcription polymerase chain reaction tests, which can detect genetic material from SARS-CoV-2 and quantify the number of viral particles in each milliliter of sample. Researchers led by Akiko Iwasaki, an immunologist at Yale, compared viral loads in saliva and NP swabs from 154 patients and 109 people without the virus. They divided the patients into groups that had low, medium, and high viral loads as determined by both types of test. Then they compared those results with the severity of symptoms the patients developed later. They found that patients who developed severe disease, were hospitalized, or died were more likely to have had high virus loads in their saliva tests, but not in their NP swabs. Viral load in both saliva and nasal mucus declined over time in patients who recovered, but not in those who died. When Iwasaki and her colleagues reviewed patients’ electronic medical records for markers of disease in the blood, they found that high saliva viral loads correlated with high levels of immune signals such as cytokines and chemokines, nonspecific molecules that ramp up in response to viral infections and have been linked to tissue damage. People with more virus in their saliva also gradually lost certain cells that mount an immune response against viral targets, had lower levels of antibodies targeting the spike protein that the virus uses to enter cells, and were slower to develop the strong immune response needed to knock down the virus in cases where they recovered. The team’s results appeared on 10 January in a preprint that has not been peer reviewed. Iwasaki and her colleagues argue that saliva may be a better predictor of disease outcome than nasal mucus because the latter comes from the upper respiratory tract, whereas severe disease is associated with damage deep in the lungs. “Saliva may better represent what is going on in the lower respiratory tract,” Iwasaki says, because cilia lining the respiratory tract naturally move mucus up from the lungs into the throat, where it mixes with saliva; coughs have the same effect. The results don’t have enough statistical power to reveal how much more likely a person with a high saliva viral load is to develop severe COVID-19, Iwasaki says. She is also eager for other groups to replicate the results, especially because efforts to link high NP swab viral loads with disease progression have had mixed results. If other research confirms the finding, “it would clear away a lot of the fog” around this disease, Crotty says. Monica Gandhi, an infectious disease expert at the University of California, San Francisco, adds that if saliva tests are predictive, they could help doctors identify patients to treat early with either antibodies to reduce viral load or steroids to tamp down overactive nonspecific immune responses. Saliva tests are cheaper and easier than NP tests, but much less widely available. So confirmation of the new results could bolster efforts to make saliva tests more readily available, says Sri Kosuri, CEO of Octant, Inc., a biotech company. “If this study happened in March, we’d be talking about whether we should be doing NP testing at all,” Kosuri says. Preprint available in medRxiv (Jan. 10, 2021): https://doi.org/10.1101/2021.01.04.21249236
Via Juan Lama
Mucosal Immunity: Feces and Covid-19
Mucosal Immunity : Feces and Covid-19 Surprisingly, although detection of virus in feces has been reported since the beginning of pandemy in China, mainly in children, the number of published papers focusing on this topic remain very low (# 250 papers in one year of pandemy, i.e 0,5% of Sars-Cov-2 published papers, more than 55000 according to PuBMed). Applications in diagnosis, epidemiology and follow-up of individual patients are almost inexistent. However, detection of viral material in effluents raised interest in published papers and are regularly reported in media. This short opinion paper wants to address this issue and stimulate researchers and diagnostic companies developping clinical research projects in this direction. If you consider results from search in PubMed , most papers (46%) originate from China some papers (10%) relate to animal coronavirus infection, and at least 15% report on sewage analysis. Papers reporting data on human diagnostic detection of virus in feces represent at most 50% of this collection. Clinically, the gastrointestinal tract is affected by COVID-19, gastrointestinal symptoms ranging from 7 to 60%, including diarrhea, nausea/vomiting, anorexia and abdominal pain. Biologically, presence of viral RNA in stools is rarely tested, but positive up to 50%. Virus was detected later than in sputum and respiratory secretions up to several weeks after clinical symptoms. RNA material is detected with frequently dynamic intra-host variations during convalescence, and sometimes probably viable virus potentially infectious. In the context, some chinese authors recommend including feces analysis particularly for discharge criteria Epidemiological and environment data were established through detection of virus RNA in sewage and wastewaters and also in solid fraction. These tests appear efficient for detecting new waves of cases and are apparently widely in use in Europe. Risk of transmission should be higher in developing countries where sanitation remains poor but data are lacking from such countries. During the past few weeks, China has been also focusing on potential dangerosity of foreign frozen foods and reported detection of viral RNA on various imported food, from diverse countries. Infectivity has not been demonstrated, As stated in a recent chinese paper, « … ignoring potential faecal transmission and the gastrointestinal involvement of SARS-CoV-2 may result in mistakes in attempts to control the pandemic. » Indeed, in recent control measures trying to limit contaminations in Beijing, ar least 3 fecal analysis became mandatory beginning of January.
The mucosal immune system is the largest component of the entire immune system, having evolved to provide protection at the main sites of infectious threat: the mucosae. As SARS-CoV-2 initially infects the upper respiratory tract, its first interactions with the immune system must occur predominantly at the respiratory mucosal surfaces, during both inductive and effector phases of the response. However, almost all studies of the immune response in COVID-19 have focused exclusively on serum antibodies and systemic cell-mediated immunity including innate responses. This article proposes that there is a significant role for mucosal immunity and for secretory as well as circulating IgA antibodies in COVID-19, and that it is important to elucidate this in order to comprehend especially the asymptomatic and mild states of the infection, which appear to account for the majority of cases. Moreover, it is possible that mucosal immunity can be exploited for beneficial diagnostic, therapeutic, or prophylactic purposes.
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
The Coronavirus Disease 2019 pandemic has made deployment of an effective vaccine a global health priority.We evaluated the protective activity of a …...
medRxiv - The Preprint Server for Health Sciences
olfactory epithelium and Covid-19 by sciencemag
L’infection par le SARS-CoV-2 s’accompagne parfois d’une perte d’odorat. La bonne nouvelle ? Celui-ci devrait très probablement être de retour une fois l’infection terminée. Plus ou moins vite.
virus in feces and on surfaces
|
One ‘superspreader’ with Omicron shed three times as much viral RNA as those with Alpha or Delta. People infected with the highly transmissible Alpha, Delta and Omicron variants of SARS-CoV-2 spew out higher amounts of virus than do those infected with other variants, according to a study1. Furthermore, individuals who contract COVID-19 after vaccination, and even after a booster dose, still shed virus into the air. The work was posted on the medRxiv preprint server on 29 July. It has not yet been peer reviewed. “This research showed that all three of those variants that have won the infection race … come out of the body more efficiently when people talk or shout than the earliest strains of the coronavirus,” says John Volckens, a public-health engineer at Colorado State University in Fort Collins. Study co-author Kristen Coleman, who researches emerging infectious diseases at the University of Maryland in College Park, says this means that people should be “pushing governments to invest in improving indoor air quality by improving ventilation and filtration systems”. Breathe out For the study, Coleman and her colleagues recruited 93 people between mid-2020 and early 2022 who were infected with SARS-CoV-2. Participants’ infections were caused by strains including the Alpha variant, which emerged in late 2020, and the later Delta and Omicron variants. All participants with the latter two strains had been fully vaccinated before catching the virus. The infected people faced into a cone-shaped apparatus and sang and shouted — with inevitable coughs and sneezes in between — for 30 minutes, while an attached machine collected the particles they exhaled. The device, called a Gesundheit-II, separated out the fine ‘aerosol’ droplets measuring 5 micrometres or less in diameter, which can linger in the air and leak through cloth and surgical masks. The team found that participants infected with the Alpha, Delta and Omicron variants emitted significantly more viral RNA when exhaling than did people infected with other variants. These include ancestral variants, such as the one first detected in Wuhan, China, and those not associated with increased transmissibility — such as Gamma, which arose in late 2020. For participants with Delta and Omicron, their fine aerosol contained on average five times the amount of virus that was detected in their larger, coarse aerosol. The team also seeded cells in the laboratory with aerosol samples and found that four samples, each from a participant with either Delta or Omicron, infected the cells. Shed virus is not always infectious, says study co-author Jianyu Lai, an epidemiologist at the University of Maryland, and the samples’ ability to infect laboratory cells means that viral RNA in exhaled aerosols can spread the disease. Malin Alsved, an aerosol technology scientist at Lund University in Sweden, says: “I’m bit concerned that they mix all the respiratory [aerosols] — they have breathing, talking, speaking, screaming, coughing, even sneezing in the sample.” Coleman responds that the team combined respiratory samples to mimic a real-life scenario such as being in a restaurant. Going viral The study also highlights variation between individuals in the amounts of exhaled virus, which ranged from non-detectable levels to those associated with ‘superspreaders’. One Omicron-infected participant, for example, shed 1,000 times as much viral RNA through fine aerosol as the maximum level observed in those with Alpha or Delta. The researchers say that the root of these discrepancies remains a mystery but could be related to biological factors such as a person’s age. Behaviour might play a part, too: the study’s superspreader coughed more frequently than others. If new variants are more prone to superspreading, that might drive them to dominate COVID-19 cases. The team notes that people infected with SARS-CoV-2 exhale much lower amounts of viral RNA than do people infected with influenza, a comparable airborne disease. This suggests that SARS-CoV-2 could spin off variants that transmit even more virus. “That is something to be concerned about,” says Alsved. Published in Nature (August 17, 2022): https://doi.org/10.1038/d41586-022-02202-z
Via Juan Lama
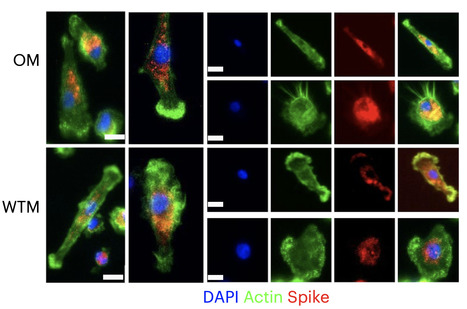
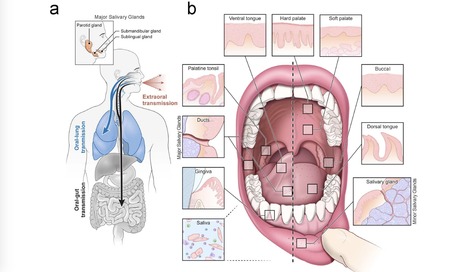
Despite signs of infection—including taste loss, dry mouth and mucosal lesions such as ulcerations, enanthema and macules—the involvement of the oral cavity in coronavirus disease 2019 (COVID-19) is poorly understood. To address this, we generated and analyzed two single-cell RNA sequencing datasets of the human minor salivary glands and gingiva (9 samples, 13,824 cells), identifying 50 cell clusters. Using integrated cell normalization and annotation, we classified 34 unique cell subpopulations between glands and gingiva. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral entry factors such as ACE2 and TMPRSS members were broadly enriched in epithelial cells of the glands and oral mucosae. Using orthogonal RNA and protein expression assessments, we confirmed SARS-CoV-2 infection in the glands and mucosae. Saliva from SARS-CoV-2-infected individuals harbored epithelial cells exhibiting ACE2 and TMPRSS expression and sustained SARS-CoV-2 infection. Acellular and cellular salivary fractions from asymptomatic individuals were found to transmit SARS-CoV-2 ex vivo. Matched nasopharyngeal and saliva samples displayed distinct viral shedding dynamics, and salivary viral burden correlated with COVID-19 symptoms, including taste loss. Upon recovery, this asymptomatic cohort exhibited sustained salivary IgG antibodies against SARS-CoV-2. Collectively, these data show that the oral cavity is an important site for SARS-CoV-2 infection and implicate saliva as a potential route of SARS-CoV-2 transmission. Published in Nature Medicine (March 25, 2021): https://doi.org/10.1038/s41591-021-01296-8
Via Juan Lama
medRxiv - The Preprint Server for Health Sciences
It has been reported that SARS-CoV-2 may use ACE2 as a receptor to gain entry into human cells, in a way similar to that of SARS-CoV. Analyzing the distribution and expression level of ACE2 may therefore help reveal underlying mechanisms of viral susceptibility ...
The distal lung contains terminal bronchioles and alveoli that facilitate gas exchange. Three-dimensional in vitro human distal lung culture systems would strongly facilitate investigation of pathologies including interstitial lung disease, cancer, and SARS-CoV-2-associated COVID-19 pneumonia. We generated long-term feeder-free, chemically defined culture of distal lung progenitors as organoids derived from single adult human alveolar epithelial type II (AT2) or KRT5+ basal cells. AT2 organoids exhibited AT1 transdifferentiation potential while basal cell organoids developed lumens lined by differentiated club and ciliated cells. Single cell analysis of basal organoid KRT5+ cells revealed a distinct ITGA6+ITGB4+ mitotic population whose proliferation further segregated to a TNFRSF12Ahi subfraction comprising ~10% of KRT5+ basal cells, residing in clusters within terminal bronchioles and exhibiting enriched clonogenic organoid growth activity. Distal lung organoids were created with apical-out polarity to display ACE2 on the exposed external surface, facilitating SARS-CoV-2 infection of AT2 and basal cultures and identifying club cells as a novel target population. This long-term, feeder-free organoid culture of human distal lung, coupled with single cell analysis, identifies unsuspected basal cell functional heterogeneity and establishes a facile in vitro organoid model for human distal lung infections including COVID-19-associated pneumonia.
The SARS-CoV-2 immune response in human milk has not yet been examined, though protecting
infants and young children from COVID-19 is critical for limiting community transmission,
and preventing serious illness and death.
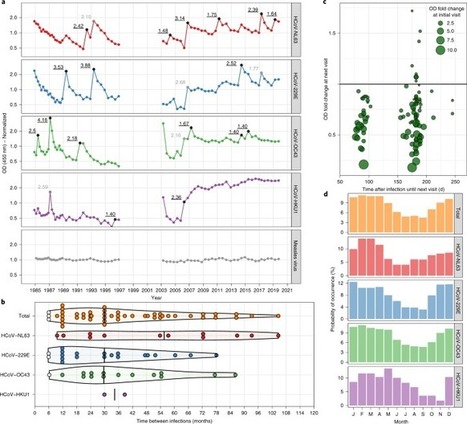
A key unsolved question in the current coronavirus disease 2019 (COVID-19) pandemic is the duration of acquired immunity. Insights from infections with the four seasonal human coronaviruses might reveal common characteristics applicable to all human coronaviruses. We monitored healthy individuals for more than 35 years and determined that reinfection with the same seasonal coronavirus occurred frequently at 12 months after infection. The durability of immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is unknown. Lessons from seasonal coronavirus infections in humans show that reinfections can occur within 12 months of initial infection, coupled with changes in levels of virus-specific antibodies.
The results of 3 new studies reveal unknown ways until now that COVID-19 spreads, leading scientists to urge the use of face coverings in public. Research from various parts of the world considered three separate factors in each study: low humidity, public restrooms and airborne dust. The implications from all three of these studies? Mask up! Low Humidity A study just published in Transboundary and Emerging Diseases confirms an earlier study conducted in the Greater Sydney, Australia area during the early stages of the pandemic that reported a link between low humidity and community spread of COVID-19. The new research led by Dr. Michael Ward in the Sydney School of Veterinary Science adds to the growing body of evidence that low humidity is a key factor in the spread of the Coronavirus. The scientists estimated that for a 1% decrease in relative humidity, COVID-19 cases increase by 7 to 8%. The same link was not found in other weather patterns such as rainfall, temperature or wind. According to Ward, dry air favors the spread of the virus: “”When the humidity is lower, the air is drier and it makes the aerosols smaller,” he said, adding that aerosols are smaller than droplets. “When you sneeze and cough those smaller infectious aerosols can stay suspended in the air for longer. That increases the exposure for other people. When the air is humid and the aerosols are larger and heavier, they fall and hit surfaces quicker.” These findings add to a growing body of research that recommends wearing a mask. Public Restrooms A second study conducted by Chinese researchers from Yangzhou University reported that flushing a public restroom toilet or urinal can release clouds of virus-laden aerosols that can be inhaled, posing a serious public health challenge. The scientists simulated and tracked virus-laden particle movements when toilets and urinals were flushed. They discovered that flushing involves an interaction between gas and liquid, resulting in a large spread of aerosol particles. The disturbing results revealed that the trajectory of the particles ejected from flushing showed that more than 57% of the particles traveled away from the urinal. The researchers further point out that when men use urinals in a public restroom, these tiny particles can reach their thigh within 5.5 seconds when compared to the toilet flush, which takes 35 seconds to reach slightly higher. “From our work, it can be inferred that urnial flushing indeed promotes the spread of bacteria and viruses,” said Xiang-Dong Liu, one of the study’s authors. “Wearing a mask should be mandatory within public restrooms during the pandemic, and anti-diffusion improvements are urgently needed to prevent the spread of COVID-19.” Airborne Dust A third study from the University of California, Davis and the Icahn School of Medicine at Mt. Sinai reported that influenza viruses can spread through the air on dust, fibers and other microscopic particles. Until now, scientists assumed airborne transmission occurred mainly due to respiratory droplets by coughing, sneezing or talking. The scientists examined whether tiny, non-respiratory particles could carry influenza between guinea pigs. Using an automated particle sizer to count airborne particles, they found that uninfected guinea pigs give off spikes of up to 1,000 particles per second as they move around the cage. Particles given off by the animals’ breathing were at a constant but lower rate. Immune guinea pigs with influenza virus painted on their fur could transmit the virus through the air to other, susceptible guinea pigs, showing that the virus did not have to come directly from the respiratory tract to be infectious. Next the researchers tested whether microscopic fibers from an inanimate object could carry infectious viruses. They treated paper facial tissues with influenza virus, let them dry out and then crumpled them in front of the automated particle sizer. Crumpling the tissues released up to 900 particles per second in a size range that could be inhaled. They were also able to infect cells from these particles released from the virus-contaminated paper tissues. According to the researchers, the surprising findings that dust can spread viruses have obvious implications for Coronavirus transmission and use of masks for protection. Plus, they said, it opens up an entire new field of investigation on how outbreaks are interpreted. Original studies available in these sites: https://doi.org/10.1063/5.0021450 https://doi.org/10.1038/s41467-020-17888-w https://doi.org/10.1111/tbed.13766
Via Juan Lama
IgA secreting plasma cells in the lamina propria are shown to be an important source of iNOS and TNF required to maintain the homeostatic balance between intestinal microbes and the immune system. The gut contains a vast number of bacteria that are essential for the health of the organism, but it is also a rich source of lymphocytes that exist to eliminate infections. How do lymphocytes restrain themselves from attacking beneficial bacteria, yet maintain their ability to respond to true pathogens? Fritz et al. show that as B cells differentiate into plasma cells in the gut, they adopt a phenotype reminiscent of innate immune cells — inflammatory monocytes — while maintaining their ability to produce immunoglobulin. The resulting immunoglobulin-A-secreting plasma cells in the lamina propria are shown to be the main source of the antimicrobial mediators tumour necrosis factor-α and inducible nitric oxide synthase, which are required to maintain the homeostatic balance between intestinal microbes and the immune system. The largest mucosal surface in the body is in the gastrointestinal tract, a location that is heavily colonized by microbes that are normally harmless. A key mechanism required for maintaining a homeostatic balance between this microbial burden and the lymphocytes that densely populate the gastrointestinal tract is the production and transepithelial transport of poly-reactive IgA (ref. 1). Within the mucosal tissues, B cells respond to cytokines, sometimes in the absence of T-cell help, undergo class switch recombination of their immunoglobulin receptor to IgA, and differentiate to become plasma cells2. However, IgA-secreting plasma cells probably have additional attributes that are needed for coping with the tremendous bacterial load in the gastrointestinal tract. Here we report that mouse IgA+ plasma cells also produce the antimicrobial mediators tumour-necrosis factor-α (TNF-α) and inducible nitric oxide synthase (iNOS), and express many molecules that are commonly associated with monocyte/granulocytic cell types. The development of iNOS-producing IgA+ plasma cells can be recapitulated in vitro in the presence of gut stroma, and the acquisition of this multifunctional phenotype in vivo and in vitro relies on microbial co-stimulation. Deletion of TNF-α and iNOS in B-lineage cells resulted in a reduction in IgA production, altered diversification of the gut microbiota and poor clearance of a gut-tropic pathogen. These findings reveal a novel adaptation to maintaining homeostasis in the gut, and extend the repertoire of protective responses exhibited by some B-lineage cells.
Early and accurate detection is critical for preventing the spread of COVID-19 and providing appropriate care for patients. Nasopharyngeal (NP) swabs, which require inserting a long shaft into the nasal cavity to collect a sample from the back of the nose and throat, are currently the gold standard for collecting a specimen for diagnosis. But the procedure is technically challenging, often uncomfortable for patients and requires personal protective equipment that may be in short supply. Other approaches to collecting specimens—including from an oropharyngeal swab and sputum—have been tested in small studies, but there is uncertainty about which method is best for detecting the virus. In a new study published in EBioMedicine, investigators from Brigham and Women's Hospital conducted a systematic review and meta-analysis, analyzing data from more than 3,000 specimens to compare the three approaches. The team found that sputum testing detected the RNA of the virus that causes COVID-19 at significantly higher rates while oropharyngeal swab testing had lower rates. Regardless of the collection method, the earlier samples were collected after symptoms began, the higher the detection rate. "The accurate diagnosis of COVID-19 has implications for health care, return-to-work, infection control and public health," said corresponding author Jonathan Li, MD, a faculty member in the Division of Infectious Diseases at the Brigham. "Our gold standard in and out of the hospital is the nasopharyngeal swab, but there's a lot of confusion about which sampling modality is best and most sensitive. Our study shows that sputum testing resulted in significantly higher rates of SARS-CoV-2 detection and supports the use of this type of testing as a valuable method for the diagnosis and monitoring of COVID-19 patients." Li and his colleagues scoured the literature—both preprints and published papers—for studies that assessed at least two respiratory sampling sites using an NP swab, oropharyngeal swab or sputum. From more than 1,000 studies, they identified 11 that met their criteria. These studies included results from a total of 3,442 respiratory tract specimens. The team examined how often each collection method produced a positive result. For NP swabs, the rate was 54 percent; for oropharyngeal swabs, 43 percent; for sputum, 71 percent. The rate of viral detection was significantly higher in sputum than either oropharyngeal swabs or NP swabs. Detection rates were highest within one week of symptom onset for all three tests. Study Published in EbioMedicine (July 18, 2020): https://doi.org/10.1016/j.ebiom.2020.102903
Via Juan Lama
A new study has revealed that SARS-CoV-2 infects the nasal cavity to the greatest degree and infects and replicates progressively less well in cells lower in the respiratory tract. The work, including the development of key laboratory tools, has been accepted by Cell.
|


 Your new post is loading...
Your new post is loading...