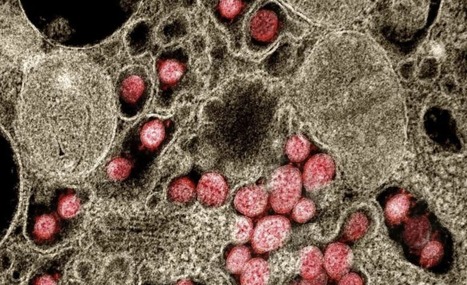
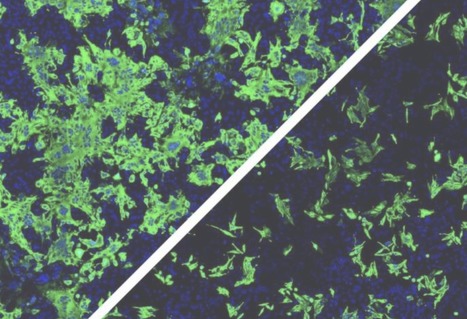
In Europe, the pandemic triggered in 2020 by the SARS-CoV-2 coronavirus is now largely under control. But why this virus is able to spread so efficiently remains unclear. A team of researchers led by Dr. Simone Backes, Dr. Gerti Beliu and Prof. Dr. Markus Sauer of the Julius Maximilians University of Würzburg (JMU) has now shown in a publication in "Angewandte Chemie" that some previous assumptions need to be reconsidered. For example, the virus does not bind with several surface proteins simultaneously to several receptors of the cell to be infected. This assumption has previously been an attempt to explain how viruses increase their infectivity. Binding to a single receptor also does not lead to the subsequent docking of further receptors to the virus. The Würzburg research group has now provided evidence that a single virus binds to a single receptor, opening the door for a highly efficient infection.
What could only be speculated about
SARS-CoV-2 carries an average of 20 - 40 spike proteins on its surface. With these, it binds to ACE2 receptors in the membrane of its target cells, for example in the nose and throat of humans. When these receptors are blocked with antibodies, the cell can no longer be infected. Making the ACE2 receptors and their interaction with the viral spike proteins visible microscopically has not been possible so far. Therefore, much was left to speculation - such as whether the viruses bind to multiple receptors with multiple spikes to facilitate entry into the cell. It was also considered that the receptors are present in the membrane in pairs or groups of three rather, so that they can bind more efficiently to the trimeric spike proteins. Or that they are only combined into such groups after binding to a spike protein. Both depend strongly on the density of the ACE2 receptors in the membrane.
Super-resolution microscopy made it clear
The Würzburg researchers wanted to elucidate this mystery: They labeled antibodies with dyes to make the receptors visible and countable. To do this, they used various cell lines that are used as model systems for SARS-CoV infection, and the single-molecule sensitive super-resolution microscopy method dSTORM, developed in Markus Sauer's research group. It turned out that Vero cells, for example, which are often used as a model for SARS-CoV-2 infection, only have one to two ACE2 receptors per square micrometre of cell membrane. This is very few: "In other membrane receptors, this number is often between 30 and 80," Sauer added. "The average distance between neighbouring ACE2 receptors is about 500 nanometres. It is thus much larger than a virus particle, which measures only 100 nanometres," says Backes. The idea that a virus particle with multiple spike proteins can bind to multiple receptors simultaneously is therefore very unlikely, she adds.
ACE2 receptors are always single
The following open question: Are the receptors also present as pairs or groups of three in the membrane? "No. They only occur there singly. And it stays that way even when a viral spike protein has bound to them," says Beliu, group leader at the Rudolf Virchow Center. For an infection, it is sufficient if a single spike binds to a single receptor. With these results, the JMU team was able to disprove many of the original hypotheses about the interaction of viral particles with multiple ACE2 receptors. It also showed that host cells with higher ACE2 expression are more easily to infect, as expected. However, the lipid composition of the membrane and other factors also influence infection efficiency.
What is next?
The JMU team wants to gather as much detailed knowledge as possible about the cell entry mechanism of coronaviruses in order to better understand the infection process. This could ultimately contribute to better prevention and the development of better drugs against COVID-19. Next, the Würzburg researchers want to analyse the entry mechanism with high-resolution light sheet microscopy.
Published March 2023:
https://doi.org/10.1002/anie.202300821



 Your new post is loading...
Your new post is loading...