In modern agricultural systems, high crop yields depend on the high input of orthophosphate-based fertilizers. Owing to the looming scarcity of rock phosphate in the near future, utilization of arbuscular mycorrhizal (AM) symbiosis for the efficient uptake of phosphorus is a necessary approach to maintain crop yield. Plant responsiveness to AM fungi is assessed based on the difference in biomass between plants with and without AM fungal inoculation. Soybean is a plant which establishes both mycorrhizal and rhizobial symbiosis. Therefore, it is difficult to eliminate the symbiotic effects of rhizobial nitrogen fixation during long-term cultivation until soybean harvest. In this study, Bradyrhizobium diazoefficiens USDA110 was inoculated at the same time as soybean seeds were sown, and soybean was grown under Rhizophagus irregularis inoculated and non-inoculated conditions to verify whether the mycorrhizal inoculation effect was observed at the yield level. In 2013, two soybean cultivars Tachinagaha and Williams 82, were grown at five levels of phosphate availability (0, 5, 10, 20, and 40 kg P 10a−1) in an open greenhouse in Tsukuba, Japan. Among the several growth indices analyzed, seed weight per plant was selected as a reliable indicator of plant response. Mycorrhizal inoculation showed a positive effect on soybean under 20 kg P 10a−1 phosphorus fertilization for Williams 82, while Tachinagaha showed almost no effect. This trend was replicated in 2014 and 2016, indicating that Tachinagaha has a lower AM responsiveness than Williams 82.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account

| Tags |
|---|
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
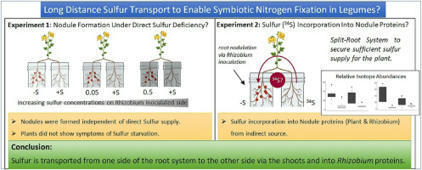
Sulfur is an essential nutrient for all plants, but also crucial for the nitrogen fixing symbiosis between legumes and rhizobia. Sulfur limitation can hamper nodule development and functioning. Until now, it remained unclear whether sulfate uptake into nodules is local or mainly systemic via the roots, and if long-distance transport from shoots to roots and into nodules occurs.
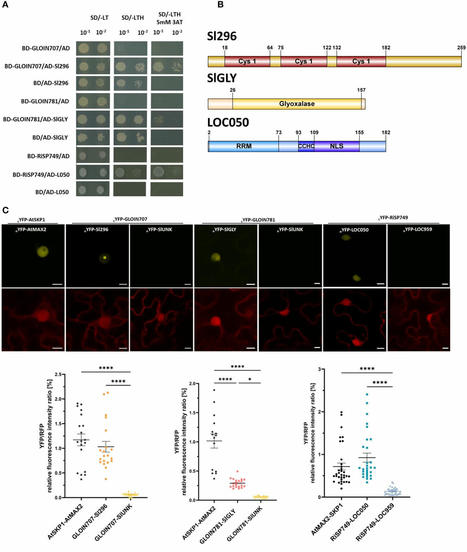
Arbuscular mycorrhizal fungi (AMF) are obligate symbionts that interact with the roots of most land plants. The genome of the AMF model species Rhizophagus irregularis contains hundreds of predicted small effector proteins that are secreted extracellularly but also into the plant cells to suppress plant immunity and modify plant physiology to establish a niche for growth. Here, we investigated the role of four nuclear-localized putative effectors, i.e., GLOIN707, GLOIN781, GLOIN261, and RiSP749, in mycorrhization and plant growth. We initially intended to execute the functional studies in Solanum lycopersicum, a host plant of economic interest not previously used for AMF effector biology, but extended our studies to the model host Medicago truncatula as well as the non-host Arabidopsis thaliana because of the technical advantages of working with these models. Furthermore, for three effectors, the implementation of reverse genetic tools, yeast two-hybrid screening and whole-genome transcriptome analysis revealed potential host plant nuclear targets and the downstream triggered transcriptional responses. We identified and validated a host protein interactors participating in mycorrhization in the host.S. lycopersicum and demonstrated by transcriptomics the effectors possible involvement in different molecular processes, i.e., the regulation of DNA replication, methylglyoxal detoxification, and RNA splicing. We conclude that R. irregularis nuclear-localized effector proteins may act on different pathways to modulate symbiosis and plant physiology and discuss the pros and cons of the tools used.
Arbuscular mycorrhizal (AM) fungi engage with land plants in a widespread, mutualistic endosymbiosis which provides their hosts with increased access to nutrients and enhanced biotic and abiotic stress resistance. The potential for reducing fertiliser use and improving crop resilience has resulted in rapidly increasing scientific interest. Microscopic quantification of the level of AM colonisation is of fundamental importance to this research, however the methods for recording and processing these data are time-consuming and tedious. In order to streamline these processes, we have developed AMScorer, an easy-to-use Excel spreadsheet, which enables the user to record data rapidly during from microscopy-based assays, and instantly performs the subsequent data processing steps. In our hands, AMScorer has more than halved the time required for data collection compared to paper-based methods. Subsequently, we developed AMReader, a user-friendly R package, which enables easy visualisation and statistical analyses of data from AMScorer. These tools require limited skills in Excel and R, and can accelerate research into AM symbioses, help researchers with variable resources to conduct research, and facilitate the storage and sharing of data from AM colonisation assays. They are available for download at https://github.com/EJarrattBarnham/AMReader, along with an extensive user manual.
Jean-Michel Ané's insight:
I look forward to seeing this tool
From
idus
Rhizobia are Gram negative bacteria that establish symbiosis with legumes and carry out the biological fixation of N 2 facilitating its assimilat ion by the plants , thus being considered an alternative to chemical fertilizers. Therefore, it is of great interest to understand all the mechanisms underlying this symbiosis. The Type VI Secretion System (T6SS) enable s bacteria to translocate effector protei ns to other bacteria or eukaryotic cells . T his system is present in Sinorhizobium fredii USDA257, a model organism used to study plant microbe interactions. However, its role remains practically unknown In this work we seek to discern th e regulation of this system in Sinorhizobium fredii USDA257 via studying the implic a tion of s ymbiotic regulator genes such as n odD2 , n olR , s yrM and m ucR . Furthermore , the d etermination of inducing and repressing media for this system is key to carry out further research towards a better understanding of the system. W e found that the T6SS is activated the most in Minimum Medium, compared to other culture media. In this medium, both gene expression and protein secretion of this system were activated to a high extent. Oppositely , in Tryptone Yeast medium the T6SS was in duced the least We aimed to obtain non functional gene mutants to test the effect of the regulators on the expression of the T6SS in both inducing and repressing conditions , obtaining simple recombinant mutants of syrM and mucR . We expect our assays to be a starting point for more advanced research on this topic, since the understudied T6SS is likely to have the potential of improv ing rhizobia legume symbiosis.
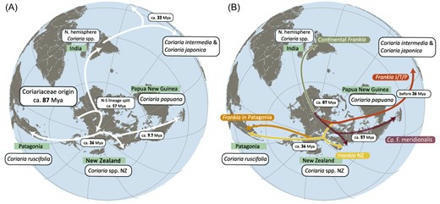
Coriariaceae are a small plant family of 14–17 species and subspecies that currently have a global but disjunct distribution. All species can form root nodules in symbiosis with diazotrophic Frankia cluster-2 strains, which form the earliest divergent symbiotic clade within this bacterial genus. Studies on Frankia cluster-2 mostly have focused on strains occurring in the northern hemisphere. Except for one strain from Papua New Guinea, namely Candidatus Frankia meridionalis Cppng1, no complete genome of Frankia associated with Coriaria occurring in the southern hemisphere has been published thus far, yet the majority of the Coriariaceae species occur here. We present field sampling data of novel Frankia cluster-2 strains, representing two novel species, which are associated with Coriaria arborea and Coriaria sarmentosa in New Zealand, and with Coriaria ruscifolia in Patagonia (Argentina), in addition to identifying Ca. F. meridionalis present in New Zealand. The novel Frankia species were found to be closely related to both Ca. F. meridionalis, and a Frankia species occurring in the Philippines, Taiwan, and Japan. Our data suggest that the different Frankia cluster-2 species diverged early after becoming symbiotic circa 100 million years ago.
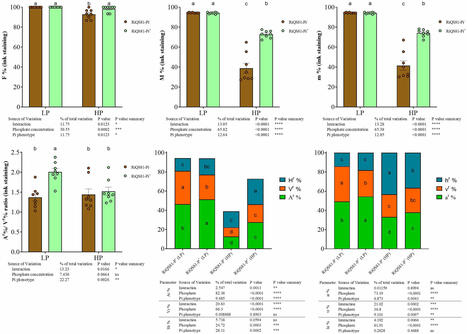
Arbuscular mycorrhizal fungi (AMF) play a crucial role in enhancing plant growth, but their use in agriculture is limited due to several constraints. Elevated soil phosphate levels resulting from fertilization practices strongly inhibit fungal development and reduce mycorrhizal growth response. Here, we investigated the possibility of adapting Rhizoglomus irregulare to high phosphate (Pi) levels to improve its tolerance. A fungal inoculum was produced through multiple generations in the presence of elevated Pi and used to inoculate melon plants grown under low and high phosphate conditions. Our results revealed distinct phenotypic and transcriptomic profiles between the adapted and non-adapted Rhizoglomus irregulare. The Pi adapted phenotype led to enhanced root colonization under high Pi conditions, increased vesicle abundance, and higher plant biomass at both phosphate levels. Additionally, the adaptation status influenced the expression of several genes involved in Pi uptake, Pi signaling, and mitochondrial respiration in both symbiotic partners. While the underlying mechanisms of the adaptation process require further investigation, our study raises intriguing questions. Do naturally occurring phosphate-tolerant AMF already exist? How might the production and use of artificially produced inocula bias our understanding? Our findings shed light on the adaptive capacities of Glomeromycota and challenge previous models suggesting that plants control mycorrhizal fungal growth. Moreover, our work pave the way for the development of innovative biotechnological tools to enhance the efficacy of mycorrhizal inoculum products under practical conditions with high phosphate fertilization.
View of the soil mycorrhizal community in a hypothetical forest dominated by oak species associated with different types of mycorrhizal fungi that form separate underground networks (MN) that may possibly be used to transport amino acids, carbohydrates, fatty acids, signaling microRNAs or peptides and hormones. Image courtesy of Florian Gadenne (Martin & van der Heijden et al., pp. 1486–1506).
Jean-Michel Ané's insight:
Awesome collection of articles in @NewPhyt
From
www
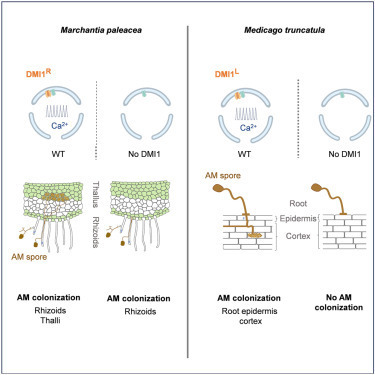
The ability of fungi to establish mycorrhizal associations with plants and enhance the acquisition of mineral nutrients stands out as a key feature of terrestrial life. Evidence indicates that arbuscular mycorrhizal (AM) association is a trait present in the common ancestor of land plants,1,2,3,4 suggesting that AM symbiosis was an important adaptation for plants in terrestrial environments.5 The activation of nuclear calcium signaling in roots is essential for AM within flowering plants.6 Given that the earliest land plants lacked roots, whether nuclear calcium signals are required for AM in non-flowering plants is unknown. To address this question, we explored the functional conservation of symbiont-induced nuclear calcium signals between the liverwort Marchantia paleacea and the legume Medicago truncatula. In M. paleacea, AM fungi penetrate the rhizoids and form arbuscules in the thalli.7 Here, we demonstrate that AM germinating spore exudate (GSE) activates nuclear calcium signals in the rhizoids of M. paleacea and that this activation is dependent on the nuclear-localized ion channel DOES NOT MAKE INFECTIONS 1 (MpaDMI1). However, unlike flowering plants, MpaDMI1-mediated calcium signaling is only required for the thalli colonization but not for the AM penetration within rhizoids. We further demonstrate that the mechanism of regulation of DMI1 has diverged between M. paleacea and M. truncatula, including a key amino acid residue essential to sustain DMI1 in an inactive state. Our study reveals functional evolution of nuclear calcium signaling between liverworts and flowering plants and opens new avenues of research into the mechanism of endosymbiosis signaling.
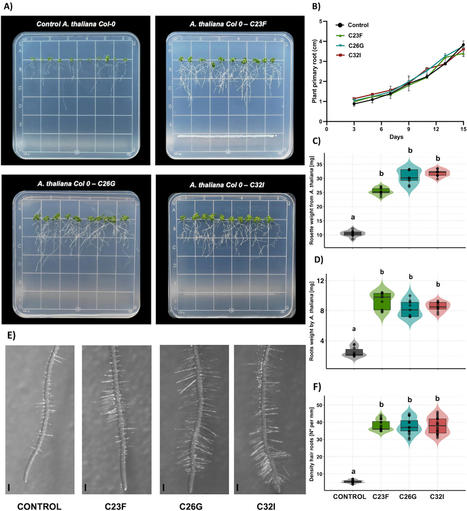
Plants and microorganisms establish beneficial associations that can improve their development and growth. Recently, it has been demonstrated that bacteria isolated from the skin of amphibians can contribute to plant growth and defense. However, the molecular mechanisms involved in the beneficial effect for the host are still unclear. In this work, we explored whether bacteria isolated from three tropical frogs species can contribute to plant growth. After a wide screening, we identified three bacterial strains with high biostimulant potential, capable of modifying the root structure of Arabidopsis thaliana plants. In addition, applying individual bacterial cultures to Solanum lycopersicum plants induced an increase in their growth. To understand the effect that these microorganisms have over the host plant, we analysed the transcriptomic profile of A. thaliana during the interaction with the C32I bacterium, demonstrating that the presence of the bacteria elicits a transcriptional response associated to plant hormone biosynthesis. Our results show that amphibian skin bacteria can function as biostimulants to improve agricultural crops growth and development by modifying the plant transcriptomic responses.
Jean-Michel Ané's insight:
This paper wins the "out of the box" prize for today :-) 
Julio Retamales's curator insight,
April 22, 11:33 AM
Nothing to add but just quoting Jean-Michel Ané's insight: "This paper wins the "out of the box" prize for today :-)"
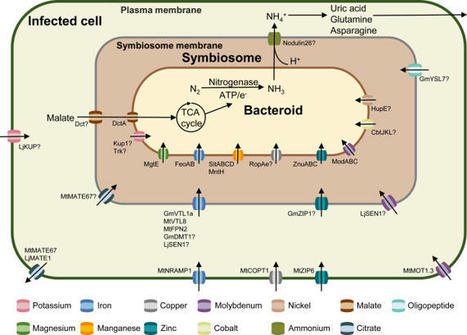
Symbiotic nitrogen fixation (SNF) facilitated by the interaction between legumes and rhizobia is a well-documented and eco-friendly alternative to chemical nitrogen fertilizers. Host plants obtain fixed nitrogen from rhizobia by providing carbon and mineral nutrients. These mineral nutrients, which are mostly in the form of metal ions, are implicated in various stages of the SNF process. This review describes the functional roles played by metal ions in nodule formation and nitrogen fixation and specifically addresses their transport mechanisms and associated transporters within root nodules. Future research directions and potential strategies for enhancing SNF efficiency are also discussed.
Symbiosis is a puzzle: seemingly in defiance of the view that biological organisms are selfish entities, nature has provided numerous examples of distantly related species “working together.” These ecological interactions have been fundamental to the emergence of Earth’s biological systems, for example, driving how eukaryotic cells evolved [1], how plants colonized the terrestrial biome [2], and how coral reef ecosystems are built [3]. Yet scratch below the surface, and collaborations between species are never fully cooperative. Many interactions remain transient maximizations of ecological, metabolic, or protective benefit. Even in long-established endosymbioses, in which a microbial symbiont lives within cells of the host, selection drives each species to evolve in directions that are often to their partner’s detriment [4]. Consequences for symbionts can include entrapment, genetic appropriation, genome reduction, loss of autonomy, expulsion, and being digested. And, from the host side, symbiosis can lead to obligate dependence on partners that are erratically present or that are mutation ridden and ecologically fragile. Such dynamics might appear to make these interactions inherently unstable, yet they arise frequently and often persist across considerable periods of time and many generations.
Jean-Michel Ané's insight:
Great editorial from Nancy Moran & Thomas Richards on #symbiosis in @PLOSBiology
A group of Gram-negative plant-associated diazotrophic bacteria belonging to the genus Nitrospirillum was investigated, including both previously characterized and newly isolated strains from diverse regions and biomes, predominantly in Brazil. Phylogenetic analysis of 16S rRNA and recA genes revealed the formation of a distinct clade consisting of thirteen strains, separate from the formally recognized species N. amazonense (the closest species) and N. iridis. Comprehensive taxonomic analyses using the whole genomes of four strains (BR 11140T = AM 18T = Y-2T = DSM 2788T = ATCC 35120T, BR 11142T = AM 14T = Y-1T = DSM 2787T = ATCC 35119T, BR 11145 = CBAmC, and BR 12005) supported the division of these strains into two species: N. amazonense (BR 11142 T and BR 12005) and a newly proposed species (BR 11140 T and BR 11145), distinct from N. iridis. The phylogenomic analysis further confirmed the presence of the new Nitrospirillum species. Additionally, MALDI-TOF MS analysis of whole-cell mass spectra provided further evidence for the differentiation of the proposed Nitrospirillum species, separate from N. amazonense. Analysis of chemotaxonomy markers (i.e., genes involved in fatty acid synthesis, metabolism and elongation, phospholipid synthesis, and quinone synthesis) revealed that the new species highlights high similarity and evolutionary convergence with other Nitrospirillum species. This new species exhibited nitrogen fixation ability in vitro, it has similar NifHDK protein phylogeny position with the closest species, lacked denitrification capability, but possessed the nosZ gene, enabling N2O reduction, distinguishing it from the closest species. Despite being isolated from diverse geographic regions, soil types, and ecological niches, no significant phenotypic or physiological differences were observed between the proposed new species and N. amazonense. Based on these findings, a new species, Nitrospirillum viridazoti sp. nov., was classified, with the strain BR 11140T (DSM 2788T, ATCC 35120T) designated as the type strain.
Jean-Michel Ané's insight:
Uncanny beast |
From
www
Legumes sanction root nodules containing rhizobial strains with low nitrogen fixation rates (ineffective fixers). Pea (Pisum sativum) nodules contain both undifferentiated bacteria and terminally differentiated nitrogen-fixing bacteroids. It is critical to understand whether sanctions act on both bacteria and bacteroids. As only undifferentiated bacteria are able to regrow from senescent nodules, if sanctioning only acts on bacteroids then ineffective strains might evade sanctioning. In addition, ineffective strains could potentially evade sanctioning by entering the same nodule as an effective strain i.e., piggybacking. P. sativum was co-inoculated with pairwise combinations of three strains of rhizobia with different effectiveness, to test whether ineffective strains can evade sanctions in this way.
Jean-Michel Ané's insight:
Very interesting preprint from @UnderwoodPlants in @PooleLabOxford on sanctions agains cheaters in pea nodules
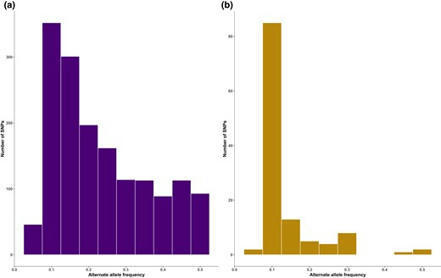
Identifying genuine polymorphic variants is a significant challenge in sequence data analysis, although detecting low-frequency variants in sequence data is essential for estimating demographic parameters and investigating genetic processes, such as selection, within populations. Arbuscular mycorrhizal (AM) fungi are multinucleate organisms, in which individual nuclei collectively operate as a population, and the extent of genetic variation across nuclei has long been an area of scientific interest. In this study, we investigated the patterns of polymorphism discovery and the alternate allele frequency distribution by comparing polymorphism discovery in 2 distinct genomic sequence datasets of the AM fungus model species, Rhizophagus irregularis strain DAOM197198. The 2 datasets used in this study are publicly available and were generated either from pooled spores and hyphae or amplified single nuclei from a single spore. We also estimated the intraorganismal variation within the DAOM197198 strain. Our results showed that the 2 datasets exhibited different frequency patterns for discovered variants. The whole-organism dataset showed a distribution spanning low-, intermediate-, and high-frequency variants, whereas the single-nucleus dataset predominantly featured low-frequency variants with smaller proportions in intermediate and high frequencies. Furthermore, single nucleotide polymorphism density estimates within both the whole organism and individual nuclei confirmed the low intraorganismal variation of the DAOM197198 strain and that most variants are rare. Our study highlights the methodological challenges associated with detecting low-frequency variants in AM fungal whole-genome sequence data and demonstrates that alternate alleles can be reliably identified in single nuclei of AM fungi.
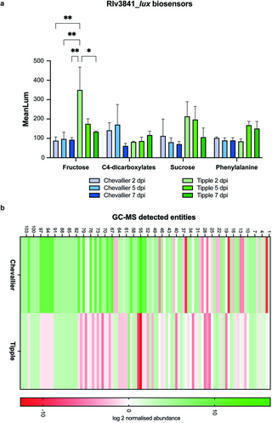
Plant-associated microbes play vital roles in promoting plant growth and health, with plants secreting root exudates into the rhizosphere to attract beneficial microbes. Exudate composition defines the nature of microbial recruitment, with different plant species attracting distinct microbiota to enable optimal adaptation to the soil environment. To more closely examine the relationship between plant genotype and microbial recruitment, we analysed the rhizosphere microbiomes of landrace (Chevallier) and modern (NFC Tipple) barley (Hordeum vulgare) cultivars. Distinct differences were observed between the plant-associated microbiomes of the 2 cultivars, with the plant-growth promoting rhizobacterial genus Pseudomonas substantially more abundant in the Tipple rhizosphere. Striking differences were also observed between the phenotypes of recruited Pseudomonas populations, alongside distinct genotypic clustering by cultivar. Cultivar-driven Pseudomonas selection was driven by root exudate composition, with the greater abundance of hexose sugars secreted from Tipple roots attracting microbes better adapted to growth on these metabolites and vice versa. Cultivar-driven selection also operates at the molecular level, with both gene expression and the abundance of ecologically relevant loci differing between Tipple and Chevallier Pseudomonas isolates. Finally, cultivar-driven selection is important for plant health, with both cultivars showing a distinct preference for microbes selected by their genetic siblings in rhizosphere transplantation assays.
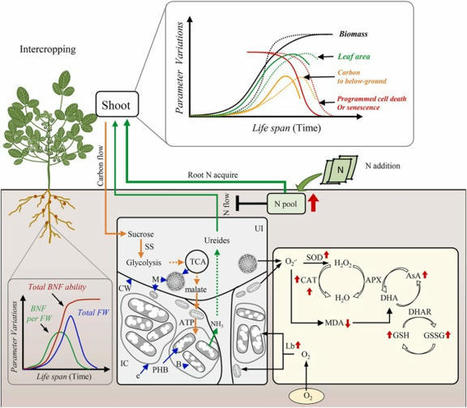
Relay intercropping soybean [Glycine max (L.) Merr.] with cereal provides an approach to increase land productivity and improve N fixation. To explore the physiological mechanisms underlying the N fixation, leaf and nodule traits, biomass and N accumulations, nodule sucrose content, paraffin section and transmission electron microscope observations, and nodule antioxidant capacity were compared in sole and relay intercropped soybean with N levels. The growth of relay intercropped soybean was suppressed during the coexistence duration, and N addition promotes leaf growth and suppressed nodule development. After maize harvest, a boosted recovery of intercropped soybean than sole cropping notably enhanced sucrose by 20.4% allocated to the nodule. Sufficient sucrose partition delayed nodule senescence by increasing antioxidant capacity in intercropped soybeans contrast in the sole cropping. The delayed nodule senescence of intercropped soybean continuously provides ureides to the aboveground. The ureide-N in total N of intercropped soybean was 37.3% notably higher than sole cropping at the R5 stage. Finally, the adverse effects of shade on soybean growth and N fixation during the coexistence duration were compensated through the recovery growth, which benefits obtaining yield advantage of the intercropped soybean.
Legume plants develop two types of root postembryonic organs, lateral roots and symbiotic nodules, using shared regulatory components. The module composed by the microRNA390, the Trans-Acting SIRNA3 (TAS3) RNA and the Auxin Response Factors (ARF)2, ARF3, and ARF4 (miR390/TAS3/ARFs) mediates the control of both lateral roots and symbiotic nodules in legumes.
In many frankia, the ability to nodulate host plants (Nod+) and fix nitrogen (Fix+) is a common strategy. However, some frankia within the Pseudofrankia genus lack one or two of these traits. This phenomenon has been consistently observed across various actinorhizal nodule isolates, displaying Nod− and/or Fix− phenotypes. Yet, the mechanisms supporting the colonization and persistence of these inefficient frankia within nodules, both with and without symbiotic strains (Nod+/Fix+), remain unclear. It is also uncertain whether these associations burden or benefit host plants. This study delves into the ecological interactions between Parafrankia EUN1f and Pseudofrankia inefficax EuI1c, isolated from Elaeagnus umbellata nodules. EUN1f (Nod+/Fix+) and EuI1c (Nod+/Fix−) display contrasting symbiotic traits. While the prediction suggests a competitive scenario, the absence of direct interaction evidence implies that the competitive advantage of EUN1f and EuI1c is likely contingent on contextual factors such as substrate availability and the specific nature of stressors in their respective habitats. In co-culture, EUN1f outperforms EuI1c, especially under specific conditions, driven by its nitrogenase activity. Iron-depleted conditions favor EUN1f, emphasizing iron’s role in microbial competition. Both strains benefit from host root exudates in pure culture, but EUN1f dominates in co-culture, enhancing its competitive traits. Nodulation experiments show that host plant preferences align with inoculum strain abundance under nitrogen-depleted conditions, while consistently favoring EUN1f in nitrogen-supplied media. This study unveils competitive dynamics and niche exclusion between EUN1f and EuI1c, suggesting that host plant may penalize less effective strains and even all strains. These findings highlight the complex interplay between strain competition and host selective pressure, warranting further research into the underlying mechanisms shaping plant–microbe–microbe interactions in diverse ecosystems.
From
www
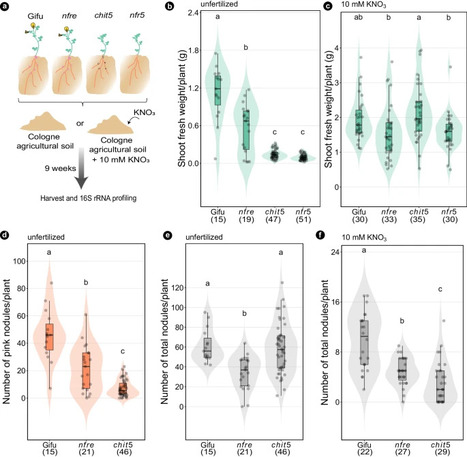
Symbiosis with soil-dwelling bacteria that fix atmospheric nitrogen allows legume plants to grow in nitrogen-depleted soil. Symbiosis impacts the assembly of root microbiota, but it is unknown how the interaction between the legume host and rhizobia impacts the remaining microbiota and whether it depends on nitrogen nutrition. Here, we use plant and bacterial mutants to address the role of Nod factor signaling on Lotus japonicus root microbiota assembly. We find that Nod factors are produced by symbionts to activate Nod factor signaling in the host and that this modulates the root exudate profile and the assembly of a symbiotic root microbiota. Lotus plants with different symbiotic abilities, grown in unfertilized or nitrate-supplemented soils, display three nitrogen-dependent nutritional states: starved, symbiotic, or inorganic. We find that root and rhizosphere microbiomes associated with these states differ in composition and connectivity, demonstrating that symbiosis and inorganic nitrogen impact the legume root microbiota differently. Finally, we demonstrate that selected bacterial genera characterizing state-dependent microbiomes have a high level of accurate prediction.
From
www
Organ-specific gene expression datasets that include hundreds to thousands of experiments allow the reconstruction of organ-level gene regulatory networks (GRNs). However, creating such datasets is greatly hampered by the requirements of extensive and tedious manual curation. Here, we trained a supervised classification model that can accurately classify the organ-of-origin for a plant transcriptome. This K-Nearest Neighbor-based multiclass classifier was used to create organ-specific gene expression datasets for the leaf, root, shoot, flower, and seed in Arabidopsis thaliana. A GRN inference approach was used to determine the: i. influential transcription factors (TFs) in each organ and, ii. most influential TFs for specific biological processes in that organ. These genome-wide, organ-delimited GRNs (OD-GRNs), recalled many known regulators of organ development and processes operating in those organs. Importantly, many previously unknown TF regulators were uncovered as potential regulators of these processes. As a proof-of-concept, we focused on experimentally validating the predicted TF regulators of lipid biosynthesis in seeds, an important food and biofuel trait. Of the top 20 predicted TFs, eight are known regulators of seed oil content, e.g., WRI1, LEC1, FUS3. Importantly, we validated our prediction of MybS2, TGA4, SPL12, AGL18, and DiV2 as regulators of seed lipid biosynthesis. We elucidated the molecular mechanism of MybS2 and show that it induces purple acid phosphatase family genes and lipid synthesis genes to enhance seed lipid content. This general approach has the potential to be extended to any species with sufficiently large gene expression datasets to find unique regulators of any trait-of-interest.
Jean-Michel Ané's insight:
Very useful. The online tool for network visualization is here: https://www.purdue.edu/hla/sites/varalalab/od-grns/
Fungi, beyond their role as saprophytes, engage in intricate relationships with plants, showcasing diverse connections ranging from mutualistic to pathogenic. We describe here an endophytic interaction employing two well-established model organisms, the grass Brachypodium distachyon and the ascomycete Neurospora crassa. Although N. crassa has been extensively investigated under laboratory conditions, its ecological characteristics remain largely unknown. In contrast, B. distachyon, a sweet grass closely related to significant crops, demonstrates remarkable ecological flexibility and participates in a variety of fungal interactions, encompassing both mutualistic and harmful associations.
Jean-Michel Ané's insight:
Surprising pre-print.. I am concerned that this interaction may not be biologically relevant. Brachypodium does not seem to like it. Was Neurospora ever isolated from Brachypoduum in the wild?
From
peerj
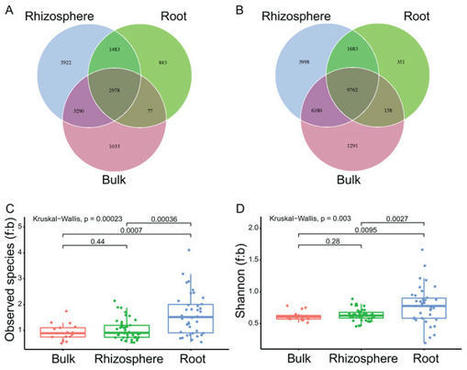
The microbial communities, inhabiting around and in plant roots, are largely influenced by the compartment effect, and in turn, promote the growth and stress resistance of the plant. However, how soil microbes are selected to the rhizosphere, and further into the roots is still not well understood. Here, we profiled the fungal, bacterial communities and their interactions in the bulk soils, rhizosphere soils and roots of eleven stress-resistant plant species after six months of growth. The results showed that the root selection (from the rhizosphere soils to the roots) was stronger than the rhizosphere selection (from the bulk soils to the rhizosphere soils) in: (1) filtering stricter on the fungal (28.5% to 40.1%) and bacterial (48.9% to 68.1%) amplicon sequence variants (ASVs), (2) depleting more shared fungal (290 to 56) and bacterial (691 to 2) ASVs measured by relative abundance, and (3) increasing the significant fungi-bacteria crosskingdom correlations (142 to 110). In addition, the root selection, but not the rhizosphere selection, significantly increased the fungi to bacteria ratios (f:b) of the observed species and shannon diversity index, indicating unbalanced effects to the fungal and bacteria communities exerted by the root selection. Based on the results of network analysis, the unbalanced root selection effects were associated with increased numbers of negative interaction (140 to 99) and crosskingdom interaction (123 to 92), suggesting the root selection intensifies the negative fungi-bacteria interactions in the roots. Our findings provide insights into the complexity of crosskingdom interactions and improve the understanding of microbiome assembly in the rhizosphere and roots.
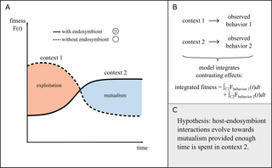
Endosymbiotic relationships are pervasive across diverse taxa of life, offering key avenues for eco-evolutionary dynamics. Although a variety of experimental and empirical frameworks have shed light on critical aspects of endosymbiosis, theoretical frameworks (mathematical models) are especially well-suited for certain tasks. Mathematical models can integrate multiple factors to determine the net outcome of endosymbiotic relationships, identify broad patterns that connect endosymbioses with other systems, simplify biological complexity, generate hypotheses for underlying mechanisms, evaluate different hypotheses, identify constraints that limit certain biological interactions, and open new lines of inquiry. This Essay highlights the utility of mathematical models in endosymbiosis research, particularly in generating relevant hypotheses. Despite their limitations, mathematical models can be used to address known unknowns and discover unknown unknowns.
Most legumes are able to develop a root nodule symbiosis in association with proteobacteria collectively called rhizobia. Among them, the tropical species Aeschynomene evenia has the remarkable property of being nodulated by photosynthetic Rhizobia without the intervention of Nod Factors (NodF). Thereby, A. evenia has emerged as a working model for investigating the NodF-independent symbiosis. Despite the availability of numerous resources and tools to study the molecular basis of this atypical symbiosis, the lack of a transformation system based on Agrobacterium tumefaciens significantly limits the range of functional approaches. In this report, we present the development of a stable genetic transformation procedure for A. evenia. We first assessed its regeneration capability and found that a combination of two growth regulators, NAA (= Naphthalene Acetic Acid) and BAP (= 6-BenzylAminoPurine) allows the induction of budding calli from epicotyls, hypocotyls and cotyledons with a high efficiency in media containing 0,5 μM NAA (up to 100% of calli with continuous stem proliferation). To optimize the generation of transgenic lines, we employed A. tumefaciens strain EHA105 harboring a binary vector carrying the hygromycin resistance gene and the mCherry fluorescent marker. Epicotyls and hypocotyls were used as the starting material for this process. We have found that one growth medium containing a combination of NAA (0,5 μM) and BAP (2,2 μM) was sufficient to induce callogenesis and A. tumefaciens strain EHA105 was sufficiently virulent to yield a high number of transformed calli. This simple and efficient method constitutes a valuable tool that will greatly facilitate the functional studies in NodF-independent symbiosis.
Jean-Michel Ané's insight:
Useful tool for an exciting emerging plant model |







Very useful study