•The quantity and activity of nutrient acquisition agents, e.g., roots, rhizobium and mycorrhizal fungi, are important for plant growth. How the invasive plants and the native legume adjust the quantity and activity of these agents in competing for soil resources remains unclear.
•The invasive plant, Xanthium strumarium, did not directly scavenge biologically fixed nitrogen (N) of a common native legume, Glycine max, but rather depleted rhizosphere soil N of the legume.
•The native legume increased its dependence on N-fixation in competition with the invasive plant, which was due to promotion of nodule activity in N-fixing by the invasive plant as well as inhibition of legume root quantity and activity.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login

Plant-Microbe Symbiosis
385.7K views |
+8 today
Follow
| Tags |
|---|
Beneficial associations between plants and microbes
Curated by
Jean-Michel Ané
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH or Gap) is a ubiquitous enzyme essential for carbon and energy metabolism in most organisms. Despite its primary role in sugar metabolism, GAPDH is recognized for its involvement in diverse cellular processes, being considered a paradigm among multifunctional/moonlighting proteins. Besides its canonical cytoplasmic location, GAPDH has been detected on cell surfaces or as a secreted protein in prokaryotes, yet little is known about its possible roles in plant symbiotic bacteria. Here we report that Rhizobium etli, a nitrogen-fixing symbiont of common beans, carries a single gap gene responsible for both GAPDH glycolytic and gluconeogenic activities. An active Gap protein is required throughout all stages of the symbiosis between R. etli and its host plant Phaseolus vulgaris. Both glycolytic and gluconeogenic Gap metabolic activities likely contribute to bacterial fitness during early and intermediate stages of the interaction, whereas GAPDH gluconeogenic activity seems critical for nodule invasion and nitrogen fixation. Although the R. etli Gap protein is secreted in a c-di-GMP related manner, no involvement of the R. etli gap gene in c-di-GMP related phenotypes, such as flocculation, biofilm formation or EPS production, was observed. Notably, the R. etli gap gene fully complemented a double gap1/gap2 mutant of Pseudomonas syringae for free life growth, albeit only partially in planta, suggesting potential specific roles for each type of Gap protein. Nevertheless, further research is required to unravel additional functions of the R. etli Gap protein beyond its essential metabolic roles.
Jean-Michel Ané's insight:
Mind the gap :-)
From
cdnsciencepub
Ammonia availability has a crucial role in agriculture as it ensures healthy plant growth and increased crop yields. Since diazotrophs are the only organisms capable of reducing dinitrogen to ammonia, they have great ecological importance and potential to mitigate the environmental and economic costs of synthetic fertilizer use. Rhizobia are especially valuable being that they can engage in nitrogen-fixing symbiotic relationships with legumes, and they demonstrate great diversity and plasticity in genomic and phenotypic traits. However, few rhizobial species have sufficient genetic tractability for synthetic biology applications. This study established a basic genetic toolbox with antibiotic resistance markers, multi-host shuttle plasmids and a streamlined protocol for biparental conjugation with Mesorhizobium and Bradyrhizobium species. We identified two repABC origins of replication from Sinorhizobium meliloti (pSymB) and Rhizobium etli (p42d) that were stable across all three strains of interest. Furthermore, the NZP2235 genome was sequenced and phylogenetic analysis determined its reclassification to Mesorhizobium huakuii. These tools will enable the use of plasmid-based strategies for more advanced genetic engineering projects and ultimately contribute towards the development of more sustainable agriculture practices by means of novel nitrogen-fixing organelles, elite bioinoculants, or symbiotic association with nonlegumes.
Jean-Michel Ané's insight:
Super useful toolkit. Thank you @jordynsmeaney, @George_diCenzo, @KarasLab and colleagues for this great resource!
Although the molecular mechanism underlying IT development remains obscure, several host players have been characterized, including VAPYRIN (VPY). The vpy mutant shows delayed initiation of ITs, often resulting in oversized MCs (Murray et al., 2011; Liu et al., 2019). VPY is also required for arbuscule development in arbuscular mycorrhiza (AM) symbiosis (Feddermann et al., 2010; Pumplin et al., 2010; Murray et al., 2011; Zhang et al., 2015; Bapaume et al., 2019; Chen et al., 2021). VPY encodes a plant-specific protein with a major sperm protein (MSP) domain and several ankyrin repeats, both of which are protein–protein interaction domains (Tarr & Scott, 2005; Li et al., 2006). In the context of nodulation, VPY interacts with LIN, a protein also essential for IT development, via its ankyrin repeats region, and is positively regulated by LIN, likely by stabilizing VPY protein by so far unknown mechanisms, in both M. truncatula and L. japonicus (Kuppusamy et al., 2004; Kiss et al., 2009; Yano et al., 2009; Liu et al., 2019; Liu et al., 2021). VPY and LIN form an ‘infectosome’ protein complex with RHIZOBIUM-DIRECTED POLAR GROWTH (RPG) and an exocyst subunit EXO70H4, regulating IT development, likely via polarized exocytosis (Arrighi et al., 2008; Liu et al., 2019; Roy et al., 2020; Wang et al., 2022; Jhu & Oldroyd, 2023; Lace et al., 2023; Li et al., 2023). The core infectosome components, VPY, LIN and RPG accumulate at the extending IT tip and form one or a few cytoplasmic puncta near the nucleus (Liu et al., 2019; Lace et al., 2023; Li et al., 2023). The functional significance and mechanism underlying the interaction of these infectosome components remain to be elucidated.
Symbiotic nitrogen fixation is an energy-intensive process, to maintain the balance between growth and nitrogen fixation, high concentrations of nitrate inhibit root nodulation. However, the precise mechanism underlying the nitrate inhibition of nodulation in soybean remains elusive. In this study, CRISPR-Cas9-mediated knockout of GmNLP1 and GmNLP4 unveiled a notable nitrate-tolerant nodulation phenotype. GmNLP1b and GmNLP4a play a significant role in the nitrate-triggered inhibition of nodulation, as the expression of nitrate-responsive genes was largely suppressed in Gmnlp1b and Gmnlp4a mutants. Furthermore, we demonstrated that GmNLP1b and GmNLP4a can bind to the promoters of GmNIC1a and GmNIC1b and activate their expression. Manipulations targeting GmNIC1a and GmNIC1b through knockdown or overexpression strategies resulted in either increased or decreased nodule number in response to nitrate. Additionally, transgenic roots that constitutively express GmNIC1a or GmNIC1b rely on both NARK and hydroxyproline O-arabinosyltransferase RDN1 to prevent the inhibitory effects imposed by nitrate on nodulation. In conclusion, this study highlights the crucial role of the GmNLP1/4-GmNIC1a/b module in mediating high nitrate-induced inhibition of nodulation.
Jean-Michel Ané's insight:
Excellent paper on the regulation of root nodulation by soil nitrogen in soybean
From
www
Legume-rhizobia root-nodule symbioses involve the recognition of rhizobial Nod factor (NF) signals by NF receptors, triggering both nodule organogenesis and rhizobial infection. RinRK1 is induced by NF signaling and is essential for infection thread (IT) formation in Lotus japonicus. However, the precise mechanism underlying this process remains unknown. Here, we show that RinRK1 interacts with the extracellular domains of NF receptors (NFR1 and NFR5) to promote their accumulation at root hair tips in response to rhizobia or NFs. Furthermore, Flotillin 1 (Flot1), a nanodomain-organizing protein, associates with the kinase domains of NFR1, NFR5 and RinRK1. RinRK1 promotes the interactions between Flot1 and NF receptors and both RinRK1 and Flot1 are necessary for the accumulation of NF receptors at root hair tips upon NF stimulation. Our study shows that RinRK1 and Flot1 play a crucial role in NF receptor complex assembly within localized plasma membrane signaling centers to promote symbiotic infection.
From
link
This 2-volume book covers the challenges, progress, and advancements towards quantitative and qualitative production of AMF for future field applications
From
www
Arbuscular mycorrhizal fungi (AMF) facilitate plant uptake of mineral nutrients, particularly phosphorus, and draw organic carbon from the plant. The ability of symbiotic AMF to utilize external non-symbiotic carbon sources remains unclear, complicating our comprehension of their ecosystem functions. Here we examine the direct absorption of exogenous 13C1-labeled myristate by symbiotic AMF and their growth responses using an in-vitro dual culture system. We also investigated the impact of exogenous myristate on the carbon‒phosphorus exchange between AMF and two different host plants in a greenhouse experiment, employing both stable isotope labeling (13CO2) and profiling of P transporter genes. Our results indicate that the extraradical hyphae of symbiotic AMF are capable of absorbing external myristate and transporting it (or its metabolic products) to intraradical structures. Myristate serves a dual function as a carbon source and signaling molecule, leading to increased intraradical and extraradical fungal biomasses, with RNA-Seq data indicating a suppressed mycorrhizal defense response as a potential mechanism. Intriguingly, exogenous myristate generally reduced the mycorrhizal phosphorus benefits for both alfalfa and rice, and decreased their carbon allocation to symbiotic AMF, likely by interfering with their normal trading mechanisms. These findings provide novel insights into the ecosystem functions and ecological applications of AMF.
Jean-Michel Ané's insight:
Greedy fungus :-)
From
academic
Bacterial persistence in the rhizosphere and colonisation of root niches are critical for the establishment of many beneficial plant-bacteria interactions including those between Rhizobium leguminosarum and its host legumes. Despite this, most studies on R. leguminosarum have focused on its symbiotic lifestyle as an endosymbiont in root nodules. Here, we use random barcode transposon sequencing (RB-TnSeq) to assay gene contributions of R. leguminosarum during competitive growth in the rhizosphere and colonisation of various plant species. This facilitated the identification of 189 genes commonly required for growth in diverse plant rhizospheres, mutation of 111 of which also affected subsequent root colonisation (rhizosphere progressive), and a further 119 genes necessary for colonisation. Common determinants reveal a need to synthesise essential compounds (amino acids, ribonucleotides, and cofactors), adapt metabolic function, respond to external stimuli, and withstand various stresses (such as changes in osmolarity). Additionally, chemotaxis and flagella-mediated motility are prerequisites for root colonisation. Many genes showed plant-specific dependencies highlighting significant adaptation to different plant species. This work provides a greater understanding of factors promoting rhizosphere fitness and root colonisation in plant-beneficial bacteria, facilitating their exploitation for agricultural benefit.
Arbuscular mycorrhizal fungi (AMF) function mostly depends on their colonization to roots. However, it has not been systematically clear how arbuscular mycorrhiza (AM) formation is affected by organic contaminants. In this study, a meta-analysis was performed to estimate the effects of three groups of organic contaminants (polycyclic aromatic hydrocarbons (PAHs), crude oil, and pesticide) in soil on mycorrhizal colonization rates (MCRs) from various moderators, such as AMF (genus, species, and inoculation mode), contaminant (level, type, and source), and experimental conditions (plant type, duration, and soil sterilization). Our results showed that all MCRs (total, vesicular, arbuscular and hyphal) significantly decreased under the stresses of three contaminants. Generally, pesticide caused larger decrease in all MCRs than PAHs and crude soil except for vesicular MCRs. Mixed AMF showed larger decrease in the total and vesicular MCRs than single AM fungus, but contrary trends in the arbuscular and hyphal MCRs. Moreover, man-made contaminant seemed to produce larger negative effects on the total, vesicular, and arbuscular MCRs than industry contaminant. The inhibitive effects of three contaminants on the total and vesicular MCRs increased with increasing experimental duration, and were larger in the non-sterilized soil than in the sterilized soil. Hyphal and spore growth were also inhibited by three contaminants in most cases. Hyphal length and spore number of single AM fungus were lower than those of mixed AMF, and pesticide produced larger inhibition on the hyphal length and spore number than PAHs and crude oil. Similar trend was found in the non-sterilized soil relative to the sterilized soil. This study indicated that AM formation and AM structure were negatively affected by three organic contaminants, whereas the inhibitive degrees varied with AMF, organic contaminant and experimental conditions.
From
link
Efficient nutrient and water acquisition from soils depends on the root system architecture (RSA). However, the genetic determinants underlying RSA in maize remain largely unexplored. In this study, we conducted a comprehensive genetic analysis for 14 shoot-borne root traits using 513 inbred lines and 800 individuals from four recombinant inbred line (RIL) populations at the mature stage across multiple field trails. Our analysis revealed substantial phenotypic variation for these 14 root traits, with a total of 389 and 344 QTLs identified through genome-wide association analysis (GWAS) and linkage analysis, respectively. These QTLs collectively explained 32.2–65.0% and 23.7–63.4% of the trait variation within each population. Several a priori candidate genes involved in auxin and cytokinin signaling pathways, such as IAA26, ARF2, LBD37 and CKX3, were found to co-localize with these loci. In addition, a total of 69 transcription factors (TFs) from 27 TF families (MYB, NAC, bZIP, bHLH and WRKY) were found for shoot-borne root traits. A total of 19 genes including PIN3, LBD15, IAA32, IAA38 and ARR12 and 19 GWAS signals were overlapped with selective sweeps. Further, significant additive effects were found for root traits, and pyramiding the favorable alleles could enhance maize root development. These findings could contribute to understand the genetic basis of root development and evolution, and provided an important genetic resource for the genetic improvement of root traits in maize.
From
www
Legumes sanction root nodules containing rhizobial strains with low nitrogen fixation rates (ineffective fixers). Pea (Pisum sativum) nodules contain both undifferentiated bacteria and terminally differentiated nitrogen-fixing bacteroids. It is critical to understand whether sanctions act on both bacteria and bacteroids. As only undifferentiated bacteria are able to regrow from senescent nodules, if sanctioning only acts on bacteroids then ineffective strains might evade sanctioning. In addition, ineffective strains could potentially evade sanctioning by entering the same nodule as an effective strain i.e., piggybacking. P. sativum was co-inoculated with pairwise combinations of three strains of rhizobia with different effectiveness, to test whether ineffective strains can evade sanctions in this way.
Jean-Michel Ané's insight:
Very interesting preprint from @UnderwoodPlants in @PooleLabOxford on sanctions agains cheaters in pea nodules
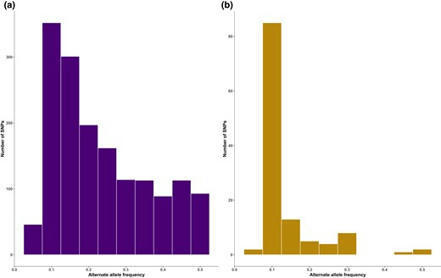
Identifying genuine polymorphic variants is a significant challenge in sequence data analysis, although detecting low-frequency variants in sequence data is essential for estimating demographic parameters and investigating genetic processes, such as selection, within populations. Arbuscular mycorrhizal (AM) fungi are multinucleate organisms, in which individual nuclei collectively operate as a population, and the extent of genetic variation across nuclei has long been an area of scientific interest. In this study, we investigated the patterns of polymorphism discovery and the alternate allele frequency distribution by comparing polymorphism discovery in 2 distinct genomic sequence datasets of the AM fungus model species, Rhizophagus irregularis strain DAOM197198. The 2 datasets used in this study are publicly available and were generated either from pooled spores and hyphae or amplified single nuclei from a single spore. We also estimated the intraorganismal variation within the DAOM197198 strain. Our results showed that the 2 datasets exhibited different frequency patterns for discovered variants. The whole-organism dataset showed a distribution spanning low-, intermediate-, and high-frequency variants, whereas the single-nucleus dataset predominantly featured low-frequency variants with smaller proportions in intermediate and high frequencies. Furthermore, single nucleotide polymorphism density estimates within both the whole organism and individual nuclei confirmed the low intraorganismal variation of the DAOM197198 strain and that most variants are rare. Our study highlights the methodological challenges associated with detecting low-frequency variants in AM fungal whole-genome sequence data and demonstrates that alternate alleles can be reliably identified in single nuclei of AM fungi. |
From
link
A novel bacterial symbiont, strain A19T, was previously isolated from a root-nodule of Aeschynomene indica and assigned to a new lineage in the photosynthetic clade of the genus Bradyrhizobium. Here data are presented for the detailed genomic and taxonomic analyses of novel strain A19T. Emphasis is placed on the analysis of genes of practical or ecological significance (photosynthesis, nitrous oxide reductase and nitrogen fixation genes). Phylogenomic analysis of whole genome sequences as well as 50 single-copy core gene sequences placed A19T in a highly supported lineage distinct from described Bradyrhizobium species with B. oligotrophicum as the closest relative. The digital DNA-DNA hybridization and average nucleotide identity values for A19T in pair-wise comparisons with close relatives were far lower than the respective threshold values of 70% and ~ 96% for definition of species boundaries. The complete genome of A19T consists of a single 8.44 Mbp chromosome and contains a photosynthesis gene cluster, nitrogen-fixation genes and genes encoding a complete denitrifying enzyme system including nitrous oxide reductase implicated in the reduction of N2O, a potent greenhouse gas, to inert dinitrogen. Nodulation and type III secretion system genes, needed for nodulation by most rhizobia, were not detected. Data for multiple phenotypic tests complemented the sequence-based analyses. Strain A19T elicits nitrogen-fixing nodules on stems and roots of A. indica plants but not on soybeans or Macroptilium atropurpureum. Based on the data presented, a new species named Bradyrhizobium ontarionense sp. nov. is proposed with strain A19T (= LMG 32638T = HAMBI 3761T) as the type strain.
A trait is an attribute influencing an organism’s performance within its environment, encompassing morphological, genetic and physiological characteristics measured at the individual or population levels (Salguero-Gómez et al. , 2018; Zhang et al. , 2023a). Understanding the ecology of species using a trait-based approach can contribute to a mechanistic explanation of processes mediated by microbes, including those that affect ecosystem functioning (Romero-Olivares et al. , 2021). This approach holds particular significance for arbuscular mycorrhizal (AM) fungi - Phylum Glomeromycota. As obligate symbionts of plants, where multiple species colonize both roots and soils in a network, predicting the functional outcomes (e.g., host growth, plant community diversity, soil characteristics) of individual AM fungal genotypes and communities within ecosystems remains challenging, despite major developments in molecular methods in the last two decades (Tisserant et al. , 2013; Montoliu-Nerin et al. , 2021). Indeed, establishing connections between AM fungal taxa and/or genotypes (e.g., within species) and their functional roles is a laborious process, which is expected to continue in the foreseeable future (Serghi et al. , 2021; Manley et al. , 2023). This is needed due to the complex links between AM fungi and functional outcomes for both hosts (e.g. , plant growth and fitness, nutrient uptake and stress tolerance) and soil functions/properties (e.g. , carbon storage, aggregate stability, biotic diversity), which appear to be highly context dependent and relatively poorly predicted by taxonomy alone (Munkvold et al. , 2004; Koch et al. , 2017; Yang et al. , 2017; Qiu et al. , 2021). However, this effort is also required because AM fungal traits have not been systematically assessed alongside with hypotheses of adaptation or with specific mechanisms in mind. For example, small-spored AM fungi may be dispersed longer distances by wind than large-spored AM fungi. It is then a reasonable hypothesis that small spore size is an adaptation for wind dispersal. One could empirically observe that small-spored AM fungi are geographically more widespread than large-spored fungi and this potential result could be viewed as evidence in support of this hypothesis. However, this finding would not necessarily prove that such dispersal difference has “functional” or “adaptive” value. Alternatively, producing small spores is a correlated response to producing many spores quickly, which itself could be an adaptive response to the likelihood of unpredictable soil disturbance such as from tillage. In this scenario, the adaptation and/or function is the production of many spores quickly to confer resistance to disturbance and then, after soil disturbance with wind erosion, small spores may also be blown farther (which may or may not improve fitness). Another example, variation in rooting depth among plants in a community may contribute to resource partitioning. But the mechanism (differential resource depletion with depth) still needs to be demonstrated separately from the trait evidence. AM fungi could contribute to equalize resource partitioning if plants with short roots associate with AM fungi that form more extensive extra-radical mycelium and vice-versa. Given these complexities, we consider the development of a robust, universally applicable trait-based framework for predicting key AM fungal functional outcomes a priority. To achieve this objective, first we must identify AM fungal traits that can be measured at morphological, physiological, and genetic levels. Second, considering the important roles of AM fungi in ecosystems, affecting host plants, soil, and the AM fungi themselves, we need to discern/hypothesize how measuring AM fungal traits impacts each of these components. For the host plant, it is crucial to consider nutrition, biomass, fitness, and survival in face of pathogens, heavy metals, salinity, drought, etc. (Delavaux et al. , 2017; Wehneret al. ). Within the soil environment, AM fungal effects on soil structure (Rillig & Mummey, 2006), nutrient cycling, carbon storage, and other members of the soil food-web are paramount (Antunes & Koyama, 2016; Frew et al. , 2021; Horsch et al. , 2023a). Regarding the fungal organism, we should focus on key aspects of their life-history strategies; reproduction and fitness, survival, dispersal, competitive ability, infectivity and abundance both within the host and soil environments (Aguilar-Trigueros et al. , 2019; Chaudharyet al. , 2020; Deveautour et al. , 2020). This requires identifying relevant proxies (sometimes termed “soft traits” in the plant ecophysiology literature) to provide easy-to-measure quantitative metrics for such complex facets of fungal life-history that can be measured across several species. Third, we need to evaluate existing standardized methods and experimental designs, or develop new ones, to measure such relevant (soft) traits, as has been done in plant ecophysiology (Pérez-Harguindeguy et al. , 2013). Measurement standardization and relevant metadata for hypothesis-driven analysis and interpretation is essential if we are to aggregate trait information from different studies into a public database, facilitating their incorporation into earth system models (e.g., (Fry et al. , 2019) and enhancing the predictability of functional processes and/or adaptations associated with AM fungi. Analogous libraries on plant traits (Kattge et al. , 2020) have proved useful to better understand trait variation along global climatic gradients (Butleret al. , 2017). Here, we aim to:
Jean-Michel Ané's insight:
A bit dry but useful
From
academic
Molecular innovations within key metabolisms can have profound impacts on element cycling and ecological distribution. Yet, much of the molecular foundations of early evolved enzymes and metabolisms are unknown. Here, we bring one such mystery to relief by probing the birth and evolution of the G-subunit protein, an integral component of certain members of the nitrogenase family, the only enzymes capable of biological nitrogen fixation. The G-subunit is a Paleoproterozoic-age orphan protein that appears more than 1 billion years after the origin of nitrogenases. We show that the G-subunit arose with novel nitrogenase metal dependence and the ecological expansion of nitrogen-fixing microbes following the transition in environmental metal availabilities and atmospheric oxygenation that began ∼2.5 billion years ago. We identify molecular features that suggest early G-subunit proteins mediated cofactor or protein interactions required for novel metal dependency, priming ancient nitrogenases and their hosts to exploit these newly diversified geochemical environments. We further examined the degree of functional specialization in G-subunit evolution with extant and ancestral homologs using laboratory reconstruction experiments. Our results indicate that permanent recruitment of the orphan protein depended on the prior establishment of conserved molecular features and showcase how contingent evolutionary novelties might shape ecologically important microbial innovations.
From
pubs
The cell plasma membrane is a two-dimensional, fluid mosaic material composed of lipids and proteins that create a semipermeable barrier defining the cell from its environment. Compared with soluble proteins, the methodologies for the structural and functional characterization of membrane proteins are challenging. An emerging tool for studies of membrane proteins in mammalian systems is a “plasma membrane on a chip,” also known as a supported lipid bilayer. Here, we create the “plant-membrane-on-a-chip,″ a supported bilayer made from the plant plasma membranes of Arabidopsis thaliana, Nicotiana benthamiana, or Zea mays. Membrane vesicles from protoplasts containing transgenic membrane proteins and their native lipids were incorporated into supported membranes in a defined orientation. Membrane vesicles fuse and orient systematically, where the cytoplasmic side of the membrane proteins faces the chip surface and constituents maintain mobility within the membrane plane. We use plant-membrane-on-a-chip to perform fluorescent imaging to examine protein–protein interactions and determine the protein subunit stoichiometry of FLOTILLINs. We report here that like the mammalian FLOTILLINs, FLOTILLINs expressed in Arabidopsis form a tetrameric complex in the plasma membrane. This plant-membrane-on-a-chip approach opens avenues to studies of membrane properties of plants, transport phenomena, biophysical processes, and protein–protein and protein–lipid interactions in a convenient, cell-free platform.
Jean-Michel Ané's insight:
Wow... I want to try that!
From
www
Finding environmentally friendly solutions for crop growth and productivity has been gaining more attention recently. This meta-analysis aims to understand the combined application of arbuscular mycorrhizal fungi (AMF) and hydroponic systems compared to AMF in conventional (soil) systems. The analysis of up-to-date studies revealed that the root colonization, calculated as the proportion of colonized root segments relative to the total root length, by AMF in conventional (soil-based) culture exceeded hydroponic (or soilless) culture systems by 16.8%. The mean root colonization by AMF was determined to be 52.3% in hydroponic systems and 61.1% in conventional systems. Within hydroponic systems, the root colonization ranged from 2% to 20% after 10 days of inoculation, and notably, it exceeded 50% after 30 to 65 days, depending on the growing substrate and species. Under hydroponics, AMF application had a higher (compared to none-inoculated) positive effect on crop biomass and yield than fruit and leaf quality (antioxidants, phenols, and sugars) as well as leaf nutrients. However, AMF do not always have the potential to improve crop growth, quality and productivity in hydroponics. Among the studies analyzed in this review, approximately 34% (no effect: 29%; negative: 5%) reported no discernible positive effect on biomass or yield, 37% (no effect: 16%; negative: 21%) on fruit or leaf quality, and 60% (no effect: 47%; negative: 13%) on nutrient levels within plant tissues. To improve the performance of AMF in hydroponic systems, the meta-analysis recommended maintaining phosphorus levels in the nutrient solution within the range of 0.15 to 15.5 mg L−1 as elevated levels (40–75 mg L−1) were found to significantly reduce AMF colonization. Additionally, it was observed that certain hydroponic techniques, such as the presence of air bubbles generated by air pumps in floating hydroponic systems (Deep Flow technique) and continuous circulation of the nutrient solution (Ebb and Flow systems), may create dynamic conditions that could potentially hinder the introduction of AMF spores into hydroponic systems and potentially compromise the integrity of the spores and hyphae.
The survival and productivity of potato plantlets transplanted from in vitro to commercial hydroponic minituber culture are crucial. Mycorrhizal fungi are an efficient tool to enhance potato nutrient uptake, survival, and productivity under these conditions. This study aimed to assess the effect of mycorrhizal fungi on improving the yield of minitubers and nutrient uptake in potatoes transferred from the in vitro environment to the greenhouse. This study was conducted in a greenhouse in Hamadan province, I.R. Iran, in 2013. The experimental design was factorial, including treatments with and without inoculation with the mycorrhizal fungus Rhizophagus irregularis and varying phosphorus (P) concentrations in Hoagland’s solution (100%, 75%, 50%, and 25% of its maximum content, i.e. 0.965, 0.482, 0.241, and 0.12 mM H2PO4-) with three replications on potato plantlets transferred from in vitro to pots. The results indicated that the highest colonization percentage was achieved with 25% P and inoculation. The highest number of minitubers (8.9), total minituber weight per plant (53.2 grams), plant height (54.6 cm), minituber (0.38%) and leaf (0.363%) P content, minituber (33 mg/kg) and leaf (65 mg/kg) manganese (Mn) content, and minituber (38 mg/kg) and leaf (65 mg/kg) zinc (Zn) content were obtained with inoculation and 75% P of Hoagland nutrient solution, which was statistically comparable to the inoculation with 100% P treatment. In summary, inoculating potato plantlets with mycorrhiza can enhance minituber yield and nutrient uptake and potentially replace approximately 50% of the P requirement in hydroponic solutions.
Jean-Michel Ané's insight:
Not so surprising...
From
www
Bacterial pathogens and nitrogen-fixing rhizobia employ type III protein secretion system (T3SS) effectors as potent tools to manipulate plant signaling pathways, thereby facilitating infection and host colonization. However, the molecular mechanisms underlying effects of rhizobial effectors on legume infection remain largely elusive. In this study, we investigated the symbiotic role of T3SS effectors in the interaction between Sinorhizobium fredii NGR234 and Lotus japonicus. Mutants deficient in the T3SS genes Tts1 or NopA showed enhanced rhizobial infection of L. japonicus roots. Further mutant analysis showed that the NopT effector negatively affects infection, while the NopM effector in the absence of NopT promotes infection. Notably, NopM interacts with the Nod factor receptors LjNFR1 and LjNFR5. NopM ubiquitinates LjNFR5 with ten ubiquitinated lysine residues in LjNFR5 identified using mass spectrometry. Expression of NopM in L. japonicus resulted in an approximately twofold increase in LjNFR5 protein levels and enhanced rhizobial infection. Our findings indicate that NopM directly interferes with the symbiotic signaling pathway through interaction and ubiquitination of Nod factor receptors, which likely benefits rhizobial infection of L. japonicus. Our research contributes to the intricate interplay between the Nod factor signaling pathway and rhizobial T3SS effectors delivered into host cells.
Jean-Michel Ané's insight:
Exciting pre-print
Agriculture is new paradigm, sustainable intensification, focuses back on beneficial soil microorganisms for their role in reducing chemical fertilizer and pesticide input and improving plant nutritional and health.More research has been done on arbuscular mycorrhizal fungi (AMF) because they form symbiotic relationships with the root systems of most land plants and make it easier for plants to absorb nutrients by creating extraradical networks of hyphae that spread out from colonized roots into the soil and serve as supplemental absorbents.Plants are protected from abiotic and biotic factors by AMF, and they also contribute in modulating the activity of antioxidant enzymes and secondary metabolites (phytochemicals), including polyphenols, anthocyanins, phytoestrogens and carotenoids.The employment of AMF symbionts to enhance the nutritional and medicinal value of active food items has come under more and more scrutiny in studies.Despite the wide physiological and genetic diversity of plant species, only a few AMFs have been used, thus limiting their full exploitation.This review study concentrates on the results of AMF on plant secondary substance biosynthesis that can improve health, as well as the standards for choosing the best symbionts to be utilized as sustainable biotechnological instruments to create food that is healthy and safe.The main objectives of the articlewas to examine the role AMF plays in improving soil's physical, biological and chemical properties. Regarding the information gaps found in this review, a discussion of potential future research is given. This will improve our understanding of AMF, encourage additional study, and aid in maintaining soil fertility.
From
academic
The biological interactions between plants and their root microbiomes are essential for plant growth, and even though plant genotype (G), soil microbiome (M), and growth conditions (environment; E) are the core factors shaping root microbiome, their relationships remain unclear. In this study, we investigated the effects of G, M, and E and their interactions on the Lotus root microbiome and plant growth using an in vitro cross-inoculation approach, which reconstructed the interactions between nine Lotus accessions and four soil microbiomes under two different environmental conditions. Results suggested that a large proportion of the root microbiome composition is determined by M and E, while G-related (G, G × M, and G × E) effects were significant but small. In contrast, the interaction between G and M had a more pronounced effect on plant shoot growth than M alone. Our findings also indicated that most microbiome variations controlled by M have little effect on plant phenotypes, whereas G × M interactions have more significant effects. Plant genotype-dependent interactions with soil microbes warrant more attention to optimize crop yield and resilience.
Extreme climatic events and related disturbances such as hurricanes are increasingly altering forest ecosystems. How these events impact forest fungal communities is poorly characterized. We examined the effect of a hurricane on mycorrhizal community structure and potential interspecific fungal interactions, inferred from OTU co-occurrences. We characterized the root fungal communities of dual-mycorrhizal plants from nine plots during two consecutive years after a category four hurricane impacted the coastal Mexican Pacific tropical forest in Jalisco. Presence-abundance matrices were used to calculate properties of mycorrhizal networks including nestedness and modularity, and to infer patterns of co-occurrence. One year after the hurricane there was a loss of links between plants and fungi. Increased network modularity and connectivity were observed after two years. We also found that disturbance changed arbuscular mycorrhizal fungal network structure more strongly than ectomycorrhizal fungal networks. Fungal guilds changed their putative interspecific interactions, from mutual exclusion in the first year to a significant increase in co-occurrence of plant pathogens, saprotrophs, and endophytes in the second year. Our results suggest that in the short term, rhizospheric interactions can be resilient to hurricanes, but fungal guilds may have divergent responses.
In modern agricultural systems, high crop yields depend on the high input of orthophosphate-based fertilizers. Owing to the looming scarcity of rock phosphate in the near future, utilization of arbuscular mycorrhizal (AM) symbiosis for the efficient uptake of phosphorus is a necessary approach to maintain crop yield. Plant responsiveness to AM fungi is assessed based on the difference in biomass between plants with and without AM fungal inoculation. Soybean is a plant which establishes both mycorrhizal and rhizobial symbiosis. Therefore, it is difficult to eliminate the symbiotic effects of rhizobial nitrogen fixation during long-term cultivation until soybean harvest. In this study, Bradyrhizobium diazoefficiens USDA110 was inoculated at the same time as soybean seeds were sown, and soybean was grown under Rhizophagus irregularis inoculated and non-inoculated conditions to verify whether the mycorrhizal inoculation effect was observed at the yield level. In 2013, two soybean cultivars Tachinagaha and Williams 82, were grown at five levels of phosphate availability (0, 5, 10, 20, and 40 kg P 10a−1) in an open greenhouse in Tsukuba, Japan. Among the several growth indices analyzed, seed weight per plant was selected as a reliable indicator of plant response. Mycorrhizal inoculation showed a positive effect on soybean under 20 kg P 10a−1 phosphorus fertilization for Williams 82, while Tachinagaha showed almost no effect. This trend was replicated in 2014 and 2016, indicating that Tachinagaha has a lower AM responsiveness than Williams 82.
Jean-Michel Ané's insight:
Very useful study
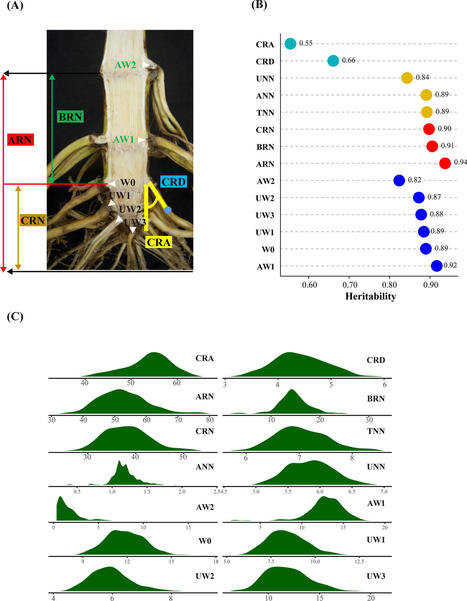
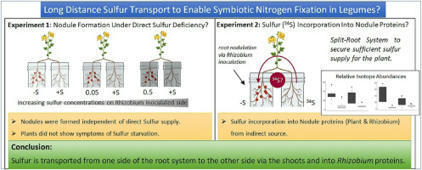
Sulfur is an essential nutrient for all plants, but also crucial for the nitrogen fixing symbiosis between legumes and rhizobia. Sulfur limitation can hamper nodule development and functioning. Until now, it remained unclear whether sulfate uptake into nodules is local or mainly systemic via the roots, and if long-distance transport from shoots to roots and into nodules occurs. |















That makes sense -